8ANI
 
 | |
8ANG
 
 | |
8ANJ
 
 | |
8ANK
 
 | |
8ANH
 
 | |
8BXV
 
 | | Crystal structure of Odorant Binding Protein 5 from Anopheles gambiae (AgamOBP5) with Thymol | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-METHYL-2-(1-METHYLETHYL)PHENOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Liggri, P.G.V, Tsitsanou, K.E, Zographos, S.E. | | Deposit date: | 2022-12-10 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The structure of AgamOBP5 in complex with the natural insect repellents Carvacrol and Thymol: Crystallographic, fluorescence and thermodynamic binding studies.
Int.J.Biol.Macromol., 237, 2023
|
|
5MUN
 
 | | Structural insight into zymogenic latency of gingipain K from Porphyromonas gingivalis. | | Descriptor: | AZIDE ION, Lys-gingipain W83 | | Authors: | Pomowski, A, Uson, I, Nowakovska, Z, Veillard, F, Sztukowska, M.N, Guevara, T, Goulas, T, Mizgalska, D, Nowak, M, Potempa, B, Huntington, J.A, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2017-01-13 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights unravel the zymogenic mechanism of the virulence factor gingipain K from Porphyromonas gingivalis, a causative agent of gum disease from the human oral microbiome.
J. Biol. Chem., 292, 2017
|
|
6GB8
 
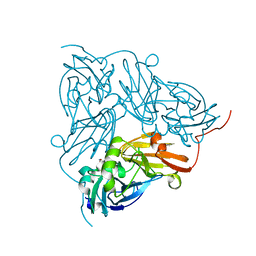 | | Copper nitrite reductase from Achromobacter cycloclastes: small cell polymorph dataset 1 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION | | Authors: | Ebrahim, A, Appleby, M.V, Axford, D, Beale, J, Moreno-Chicano, T, Sherrell, D.A, Strange, R.W, Owen, R.L, Hough, M.A. | | Deposit date: | 2018-04-13 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Resolving polymorphs and radiation-driven effects in microcrystals using fixed-target serial synchrotron crystallography.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6GBB
 
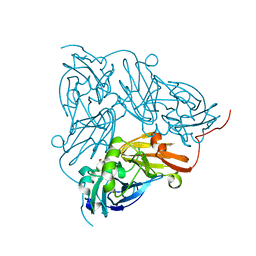 | | Copper nitrite reductase from Achromobacter cycloclastes: large cell polymorph dataset 1 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION | | Authors: | Ebrahim, A, Appleby, M.V, Axford, D, Beale, J, Moreno-Chicano, T, Sherrell, D.A, Strange, R.W, Owen, R.L, Hough, M.A. | | Deposit date: | 2018-04-13 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Resolving polymorphs and radiation-driven effects in microcrystals using fixed-target serial synchrotron crystallography.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6GCG
 
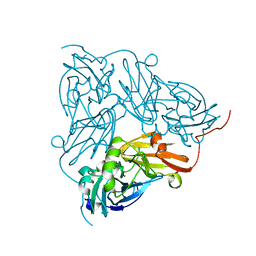 | | Copper nitrite reductase from Achromobacter cycloclastes: large polymorph dataset 15 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Ebrahim, A, Appleby, M.V, Axford, D, Beale, J, Moreno-Chicano, T, Sherrell, D.A, Strange, R.W, Owen, R.L, Hough, M.A. | | Deposit date: | 2018-04-17 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.80015242 Å) | | Cite: | Resolving polymorphs and radiation-driven effects in microcrystals using fixed-target serial synchrotron crystallography.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6J0I
 
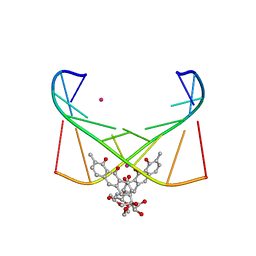 | | Structure of [Co2+-(Chromomycin A3)2]-d(TTGGCGAA)2 complex | | Descriptor: | 1,2-HYDRO-1-OXY-3,4-HYDRO-3-(1-METHOXY-2-OXY-3,4-DIHYDROXYPENTYL)-8,9-DIHYROXY-7-METHYLANTHRACENE, 2,6-dideoxy-4-O-methyl-alpha-D-galactopyranose-(1-3)-(2R,3R,6R)-6-hydroxy-2-methyltetrahydro-2H-pyran-3-yl acetate, 3-C-methyl-4-O-acetyl-alpha-L-Olivopyranose-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol, ... | | Authors: | Satange, R.B, Chuang, C.Y, Hou, M.H. | | Deposit date: | 2018-12-24 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Polymorphic G:G mismatches act as hotspots for inducing right-handed Z DNA by DNA intercalation.
Nucleic Acids Res., 47, 2019
|
|
3OW9
 
 | |
6J0H
 
 | | Crystal structure of Actinomycin D- d(TTGGCGAA) complex | | Descriptor: | Actinomycin D, DNA (5'-D(P*TP*TP*GP*GP*CP*GP*AP*A)-3'), SODIUM ION | | Authors: | Satange, R.B, Hou, M.H. | | Deposit date: | 2018-12-24 | | Release date: | 2019-07-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Polymorphic G:G mismatches act as hotspots for inducing right-handed Z DNA by DNA intercalation.
Nucleic Acids Res., 47, 2019
|
|
1P2I
 
 | | Structural consequences of accommodation of four non-cognate amino-acid residues in the S1 pocket of bovine trypsin and chymotrypsin | | Descriptor: | CALCIUM ION, Pancreatic trypsin inhibitor, SULFATE ION, ... | | Authors: | Helland, R, Czapinska, H, Leiros, I, Olufsen, M, Otlewski, J, Smalaas, A.O. | | Deposit date: | 2003-04-15 | | Release date: | 2004-04-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural consequences of accommodation of four non-cognate amino acid residues in the S1 pocket of bovine trypsin and chymotrypsin.
J.Mol.Biol., 333, 2003
|
|
1P2J
 
 | | Structural consequences of accommodation of four non-cognate amino-acid residues in the S1 pocket of bovine trypsin and chymotrypsin | | Descriptor: | CALCIUM ION, Pancreatic trypsin inhibitor, SULFATE ION, ... | | Authors: | Helland, R, Czapinska, H, Leiros, I, Olufsen, M, Otlewski, J, Smalaas, A.O. | | Deposit date: | 2003-04-15 | | Release date: | 2004-04-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural consequences of accommodation of four non-cognate amino acid residues in the S1 pocket of bovine trypsin and chymotrypsin.
J.Mol.Biol., 333, 2003
|
|
1P2K
 
 | | Structural consequences of accommodation of four non-cognate amino-acid residues in the S1 pocket of bovine trypsin and chymotrypsin | | Descriptor: | CALCIUM ION, Pancreatic trypsin inhibitor, SULFATE ION, ... | | Authors: | Helland, R, Czapinska, H, Leiros, I, Olufsen, M, Otlewski, J, Smalaas, A.O. | | Deposit date: | 2003-04-15 | | Release date: | 2004-04-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural consequences of accommodation of four non-cognate amino acid residues in the S1 pocket of bovine trypsin and chymotrypsin.
J.Mol.Biol., 333, 2003
|
|
6FHC
 
 | |
6FHD
 
 | |
2CHA
 
 | |
4EJF
 
 | |
5LYM
 
 | |
1NN6
 
 | | Human Pro-Chymase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chymase | | Authors: | Reiling, K.K, Krucinski, J, Miercke, L.J.W, Raymond, W.W, Caughey, G.H, Stroud, R.M. | | Deposit date: | 2003-01-12 | | Release date: | 2003-03-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of human pro-chymase: a model for the activating transition of granule-associated proteases.
Biochemistry, 42, 2003
|
|
3T0U
 
 | |
4N8M
 
 | | Structural polymorphism in the N-terminal oligomerization domain of NPM1 | | Descriptor: | COBALT (II) ION, Nucleophosmin | | Authors: | Mitrea, D, Royappa, G, Buljan, M, Yun, M, Pytel, N, Satumba, J, Nourse, A, Park, C, Babu, M.M, White, S.W, Kriwacki, R.W. | | Deposit date: | 2013-10-17 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structural polymorphism in the N-terminal oligomerization domain of NPM1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3SXX
 
 | |
