1GWE
 
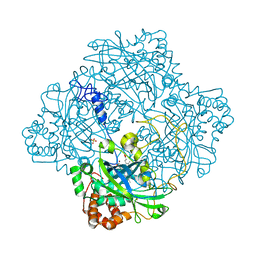 | | Atomic resolution structure of Micrococcus Lysodeikticus catalase | | Descriptor: | Catalase, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Murshudov, G.N, Grebenko, A.I, Brannigan, J.A, Antson, A.A, Barynin, V.V, Dodson, G.G, Dauter, Z, Wilson, K.S, Melik-Adamyan, W.R. | | Deposit date: | 2002-03-15 | | Release date: | 2002-04-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.88 Å) | | Cite: | The Structures of Micrococcus Lysodeikticus Catalase, its Ferryl Intermediate (Compound II) and Nadph Complex.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
5LP9
 
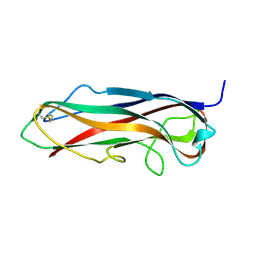 | | FimA wt from S. flexneri | | Descriptor: | Major type 1 subunit fimbrin (Pilin) | | Authors: | Zyla, D, Capitani, G, Prota, A, Glockshuber, R. | | Deposit date: | 2016-08-12 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.88626635 Å) | | Cite: | Alternative folding to a monomer or homopolymer is a common feature of the type 1 pilus subunit FimA from enteroinvasive bacteria.
J.Biol.Chem., 2019
|
|
4O6U
 
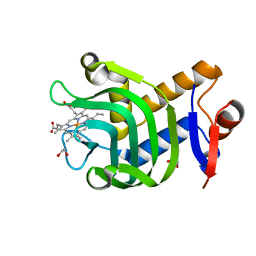 | | 0.89A resolution structure of the hemophore HasA from Pseudomonas aeruginosa (H83A mutant) | | Descriptor: | 1,2-ETHANEDIOL, HasAp, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lovell, S, Kumar, R, Battaile, K.P, Matsumura, H, Yao, H, Rodriguez, J.C, Moenne-Loccoz, P, Rivera, M. | | Deposit date: | 2013-12-23 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Replacing the Axial Ligand Tyrosine 75 or Its Hydrogen Bond Partner Histidine 83 Minimally Affects Hemin Acquisition by the Hemophore HasAp from Pseudomonas aeruginosa.
Biochemistry, 53, 2014
|
|
8GBA
 
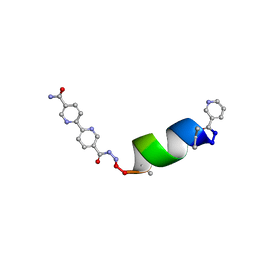 | | Porous framework formed by assembly of a bipyridyl-conjugated helical peptide | | Descriptor: | 5'-(hydrazinecarbonyl)[2,2'-bipyridine]-5-carboxamide, LEU-(AIB)-ALA-SER-LEU-ALA-CYS-(AIB)-LEU, NICOTINIC ACID | | Authors: | Hess, S.S, Nguyen, A.I. | | Deposit date: | 2023-02-24 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Noncovalent Peptide Assembly Enables Crystalline, Permutable, and Reactive Thiol Frameworks.
J.Am.Chem.Soc., 145, 2023
|
|
8GK2
 
 | | Porous framework formed by assembly of a bipyridyl-conjugated helical peptide | | Descriptor: | 5'-(hydrazinecarbonyl)[2,2'-bipyridine]-5-carboxamide, LEU-AIB-ALA-SER-LEU-ALA-CYS-AIB-LEU, NICOTINIC ACID, ... | | Authors: | Hess, S.S, Nguyen, A.I. | | Deposit date: | 2023-03-16 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Noncovalent Peptide Assembly Enables Crystalline, Permutable, and Reactive Thiol Frameworks.
J.Am.Chem.Soc., 145, 2023
|
|
4UYR
 
 | | X-ray structure of the N-terminal domain of the flocculin Flo11 from Saccharomyces cerevisiae | | Descriptor: | FLOCCULATION PROTEIN FLO11, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Veelders, M, Kraushaar, T, Brueckner, S, Rhinow, D, Moesch, H.U, Essen, L.O. | | Deposit date: | 2014-09-03 | | Release date: | 2015-08-12 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Interactions by the Fungal Flo11 Adhesin Depend on a Fibronectin Type III-Like Adhesin Domain Girdled by Aromatic Bands.
Structure, 23, 2015
|
|
3UI6
 
 | | 0.89 A resolution crystal structure of human Parvulin 14 in complex with oxidized DTT | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 4, SODIUM ION, ... | | Authors: | Mueller, J.W, Link, N.M, Matena, A, Hoppstock, L, Rueppel, A, Bayer, P, Blankenfeldt, W. | | Deposit date: | 2011-11-04 | | Release date: | 2012-11-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | 0.89 A resolution crystal structure of human Parvulin 14 in complex with oxidized DTT
To be Published
|
|
1ENN
 
 | | SOLVENT ORGANIZATION IN AN OLIGONUCLEOTIDE CRYSTAL: THE STRUCTURE OF D(GCGAATTCG)2 AT ATOMIC RESOLUTION | | Descriptor: | CHLORIDE ION, DNA (5'-D(*GP*CP*GP*AP*AP*TP*TP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Soler-Lopez, M, Malinina, L, Subirana, J.A. | | Deposit date: | 2000-03-21 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Solvent organization in an oligonucleotide crystal. The structure of d(GCGAATTCG)2 at atomic resolution.
J.Biol.Chem., 275, 2000
|
|
8R5K
 
 | |
6KFN
 
 | | Crystal structure of alginate lyase from Paenibacillus sp. str. FPU-7 | | Descriptor: | IMIDAZOLE, SODIUM ION, alginate lyase | | Authors: | Itoh, T, Nakagawa, E, Yoda, M, Nakaichi, A, Hibi, T, Kimoto, H. | | Deposit date: | 2019-07-08 | | Release date: | 2019-10-30 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Structural and biochemical characterisation of a novel alginate lyase from Paenibacillus sp. str. FPU-7.
Sci Rep, 9, 2019
|
|
7P6M
 
 | | Hydrogenated refolded hen egg-white lysozyme | | Descriptor: | ACETATE ION, Lysozyme C, NITRATE ION | | Authors: | Ramos, J, Laux, V, Haertlein, M, Forsyth, V.T, Mossou, E, Larsen, S, Langkilde, A.E. | | Deposit date: | 2021-07-16 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | The impact of folding modes and deuteration on the atomic resolution structure of hen egg-white lysozyme.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
4OY5
 
 | | 0.89 Angstrom resolution crystal structure of (Gly-Pro-Hyp)10 | | Descriptor: | Collagen | | Authors: | Suzuki, H, Mahapatra, D, Steel, P.J, Dyer, J, Dobson, R.C.J, Gerrard, J.A, Valery, C. | | Deposit date: | 2014-02-10 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Sub-angstrom structure of the collagen model peptide (GPO)10 shows a hydrated triple helix with pitch variation and two proline ring conformations
To Be Published
|
|
4UA7
 
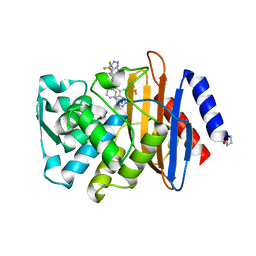 | |
4U9H
 
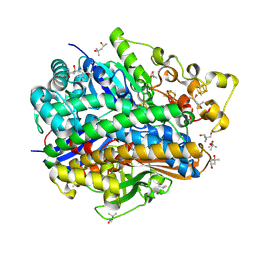 | | Ultra High Resolution Structure Of The Ni-R State Of [Nife]Hydrogenase From Desulufovibrio Vulgaris Miyazaki F | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE3-S4 CLUSTER, ... | | Authors: | Ogata, H, Nishikawa, K, Lubitz, W. | | Deposit date: | 2014-08-06 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Hydrogens detected by subatomic resolution protein crystallography in a [NiFe] hydrogenase.
Nature, 520, 2015
|
|
8JZ8
 
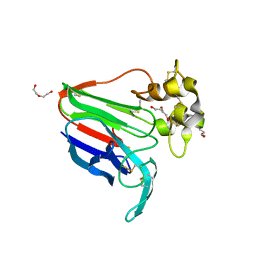 | | Subatomic structure of orthorhombic thaumatin at 0.89 Angstroms | | Descriptor: | DI(HYDROXYETHYL)ETHER, Thaumatin I | | Authors: | Masuda, T, Suzuki, M, Yamasaki, M, Mikami, B. | | Deposit date: | 2023-07-04 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Subatomic structure of orthorhombic thaumatin at 0.89 angstrom reveals that highly flexible conformations are crucial for thaumatin sweetness.
Biochem.Biophys.Res.Commun., 703, 2024
|
|
1ETL
 
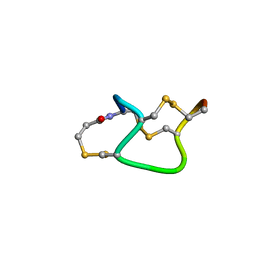 | |
1ETM
 
 | |
1SK5
 
 | |
3IP0
 
 | |
4X1A
 
 | | Lambda-[Ru(TAP)2(dppz-10,12-Me)]2+ bound to d(TCGGCGCCGA) | | Descriptor: | (10,12-dimethyldipyrido[3,2-a:2',3'-c]phenazine-kappa~2~N~4~,N~5~)[bis(pyrazino[2,3-f]quinoxaline-kappa~2~N~1~,N~10~)]ruthenium, BARIUM ION, DNA (5'-D(*TP*CP*GP*GP*CP*GP*CP*CP*GP*A)-3') | | Authors: | Hall, J.P, Cardin, C.J. | | Deposit date: | 2014-11-24 | | Release date: | 2015-05-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | The Structural Effect of Methyl Substitution on the Binding of Polypyridyl Ru-dppz Complexes to DNA
Organometallics, 2015
|
|
8PB5
 
 | | PsiM in complex with sinefungin and norbaeocystin | | Descriptor: | CHLORIDE ION, Norbaeocystin, Psilocybin synthase, ... | | Authors: | Werten, S, Hudspeth, J, Rupp, B. | | Deposit date: | 2023-06-08 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Methyl transfer in psilocybin biosynthesis.
Nat Commun, 15, 2024
|
|
1OB6
 
 | | Cephaibol B | | Descriptor: | ACETATE ION, CEPHAIBOL B, ETHANOL | | Authors: | Bunkoczi, G, Schiell, M, Vertesy, L, Sheldrick, G.M. | | Deposit date: | 2003-01-24 | | Release date: | 2003-12-11 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Crystal Structures of Cephaibols
J.Pept.Sci., 9, 2003
|
|
1HHY
 
 | | Deglucobalhimycin in complex with D-Ala-D-Ala | | Descriptor: | (2R,4S,6S)-4-azanyl-4,6-dimethyl-oxane-2,5,5-triol, D-ALANINE, DEGLUCOBALHIMYCIN, ... | | Authors: | Lehmann, C, Bunkoczi, G, Sheldrick, G.M, Vertesy, L. | | Deposit date: | 2000-12-29 | | Release date: | 2003-09-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Structures of Glycopeptide Antibiotics with Peptides that Model Bacterial Cell-Wall Precursors
J.Mol.Biol., 318, 2002
|
|
7APW
 
 | | The Fk1 domain of FKBP51 in complex with (1S,5S,6R)-10-(benzo[d]thiazol-6-ylsulfonyl)-5-(methoxymethyl)-3-(pyridin-2-ylethyl)-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,5S,6R)-10-(benzo[d]thiazol-6-ylsulfonyl)-5-(methoxymethyl)-3-(pyridin-2-ylethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Voll, A.M, Kolos, J.M, Pomplun, S, Riess, B, Purder, P, Merz, S, Bracher, A, Meyners, C, Krewald, V, Hausch, F. | | Deposit date: | 2020-10-20 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Picomolar FKBP inhibitors enabled by a single water-displacing methyl group in bicyclic [4.3.1] aza-amides.
Chem Sci, 12, 2021
|
|
1AB1
 
 | | SI FORM CRAMBIN | | Descriptor: | CRAMBIN (SER22/ILE25), ETHANOL | | Authors: | Teeter, M.M, Yamano, A. | | Deposit date: | 1997-01-31 | | Release date: | 1997-08-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Crystal structure of Ser-22/Ile-25 form crambin confirms solvent, side chain substate correlations.
J.Biol.Chem., 272, 1997
|
|
