1EB7
 
 | |
1DY8
 
 | | Inhibition of the Hepatitis C Virus NS3/4A Protease. The Crystal Structures of Two Protease-Inhibitor Complexes (inhibitor II) | | Descriptor: | N-[(benzyloxy)carbonyl]-L-isoleucyl-N-[(1R)-1-(carboxycarbonyl)-3,3-difluoropropyl]-L-leucinamide, NONSTRUCTURAL PROTEIN NS4A (P4), PROTEASE/HELICASE NS3 (P70) | | Authors: | Di Marco, S, Rizzi, M, Volpari, C, Walsh, M, Narjes, F, Colarusso, S, De Francesco, R, Matassa, V.G, Sollazzo, M. | | Deposit date: | 2000-01-31 | | Release date: | 2001-01-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibition of the Hepatitis C Virus Ns3/4A Protease the Crystal Structures of Two Protease-Inhibitor Complexes
J.Biol.Chem., 275, 2000
|
|
1ZRZ
 
 | | Crystal Structure of the Catalytic Domain of Atypical Protein Kinase C-iota | | Descriptor: | 3-{1-[3-(DIMETHYLAMINO)PROPYL]-1H-INDOL-3-YL}-4-(1H-INDOL-3-YL)-1H-PYRROLE-2,5-DIONE, Protein kinase C, iota | | Authors: | Messerschmidt, A, Macieira, S, Velarde, M, Baedeker, M, Benda, C, Jestel, A, Brandstetter, H, Neuefeind, T, Blaesse, M, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-05-23 | | Release date: | 2005-09-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Atypical Protein Kinase C-iota Reveals Interaction Mode of Phosphorylation Site in Turn Motif
J.Mol.Biol., 352, 2005
|
|
3PYE
 
 | | Mycobacterium tuberculosis 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase (IspE) in complex with CDPME | | Descriptor: | 4-DIPHOSPHOCYTIDYL-2-C-METHYL-D-ERYTHRITOL, 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase | | Authors: | Shan, S, Chen, X.H, Liu, T, Zhao, H.C, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2010-12-13 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structural Basis for anti-TB Drug Discovery targeting of 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase (IspE) from Mycobacterium tuberculosis
To be Published
|
|
1I52
 
 | | CRYSTAL STRUCTURE OF 4-DIPHOSPHOCYTIDYL-2-C-METHYLERYTHRITOL (CDP-ME) SYNTHASE (YGBP) INVOLVED IN MEVALONATE INDEPENDENT ISOPRENOID BIOSYNTHESIS | | Descriptor: | 4-DIPHOSPHOCYTIDYL-2-C-METHYLERYTHRITOL SYNTHASE, CALCIUM ION, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Richard, S.B, Bowman, M.E, Kwiatkowski, W, Kang, I, Chow, C, Lillo, M, Cane, D.E, Noel, J.P. | | Deposit date: | 2001-02-23 | | Release date: | 2001-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of 4-diphosphocytidyl-2-C- methylerythritol synthetase involved in mevalonate- independent isoprenoid biosynthesis.
Nat.Struct.Biol., 8, 2001
|
|
4RT1
 
 | | Structure of the Alg44 PilZ domain (R95A mutant) from Pseudomonas aeruginosa PAO1 in complex with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Alginate biosynthesis protein Alg44, CHLORIDE ION, ... | | Authors: | Whitfield, G.B, Whitney, J.C. | | Deposit date: | 2014-11-12 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Dimeric c-di-GMP Is Required for Post-translational Regulation of Alginate Production in Pseudomonas aeruginosa.
J.Biol.Chem., 290, 2015
|
|
1L8J
 
 | | Crystal Structure of the Endothelial Protein C Receptor and Bound Phospholipid Molecule | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endothelial protein C receptor, ... | | Authors: | Oganesyan, V, Oganesyan, N, Terzyan, S, Dongfeng, Q, Dauter, Z, Esmon, N.L, Esmon, C.T. | | Deposit date: | 2002-03-20 | | Release date: | 2002-06-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the endothelial protein C receptor and a bound phospholipid.
J.Biol.Chem., 277, 2002
|
|
1W3C
 
 | | Crystal structure of the Hepatitis C Virus NS3 Protease in complex with a peptidomimetic inhibitor | | Descriptor: | 3-({(2S)-2-[({(1R)-1-[({(1R)-1-[(R)-CARBOXY(HYDROXY)METHYL]-3,3-DIFLUOROPROPYL}AMINO)CARBONYL]-3-METHYLBUTYL}AMINO)CARB ONYL]-2,3-DIHYDRO-1H-INDOL-2-YL}METHYL)THIOPHENE-2-CARBOXYLIC ACID, 3-({(2S)-2-[({(1S)-1-[({(1S)-1-[(R)-CARBOXY(HYDROXY)METHYL]-3,3-DIFLUOROPROPYL}AMINO)CARBONYL]-3-METHYLBUTYL}AMINO)CARBONYL]-2,3-DIHYDRO-1H-INDOL-2-YL}METHYL)THIOPHENE-2-CARBOXYLIC ACID, NONSTRUCTURAL PROTEIN NS4A (P4), ... | | Authors: | Di Marco, S, Volpari, C. | | Deposit date: | 2004-07-14 | | Release date: | 2004-12-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Design and Enzyme-Bound Crystal Structure of Indoline Based Peptidomimetic Inhibitors of Hepatitis C Virus Ns3 Protease
J.Med.Chem., 47, 2004
|
|
2AWW
 
 | | Synapse associated protein 97 PDZ2 domain variant C378G with C-terminal GluR-A peptide | | Descriptor: | 18-residue C-terminal peptide from glutamate receptor, ionotropic, AMPA1, ... | | Authors: | Von Ossowski, I, Oksanen, E, Von Ossowski, L, Cai, C, Sundberg, M, Goldman, A, Keinanen, K. | | Deposit date: | 2005-09-02 | | Release date: | 2006-08-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of the second PDZ domain of SAP97 in complex with a GluR-A C-terminal peptide
Febs J., 273, 2006
|
|
1NML
 
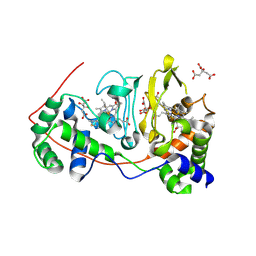 | | Di-haemic Cytochrome c Peroxidase from Pseudomonas nautica 617, form IN (pH 4.0) | | Descriptor: | CITRIC ACID, HEME C, di-haem cytochrome c peroxidase | | Authors: | Dias, J.M, Bonifacio, C, Alves, T, Pereira, A.S, Bourgeois, D, Moura, I, Romao, M.J. | | Deposit date: | 2003-01-10 | | Release date: | 2004-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the mechanism of Ca(2+) activation of the di-heme cytochrome c peroxidase from Pseudomonas nautica 617
Structure, 12, 2004
|
|
2BYE
 
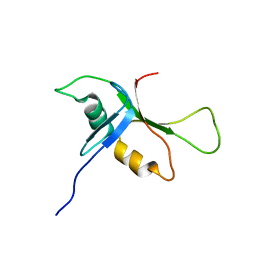 | | NMR solution structure of phospholipase c epsilon RA 1 domain | | Descriptor: | PHOSPHOLIPASE C, EPSILON 1 | | Authors: | Bunney, T.D, Harris, R, Gandarillas, N.L, Josephs, M.B, Roe, S.M, Paterson, H.F, Rodrigues-Lima, F, Esposito, D, Gieschik, P, Pearl, L.H, Driscoll, P.C, Katan, M. | | Deposit date: | 2005-08-01 | | Release date: | 2006-02-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural and Mechanistic Insights Into Ras Association Domains of Phospholipase C Epsilon.
Mol.Cell, 21, 2006
|
|
2BYF
 
 | | NMR solution structure of phospholipase c epsilon RA 2 domain | | Descriptor: | PHOSPHOLIPASE C, EPSILON 1 | | Authors: | Bunney, T.D, Harris, R, Gandarillas, N.L, Josephs, M.B, Roe, S.M, Paterson, H.F, Rodrigues-Lima, F, Esposito, D, Gieschik, P, Pearl, L.H, Driscoll, P.C, Katan, M. | | Deposit date: | 2005-08-01 | | Release date: | 2006-02-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural and Mechanistic Insights Into Ras Association Domains of Phospholipase C Epsilon.
Mol.Cell, 21, 2006
|
|
2C1U
 
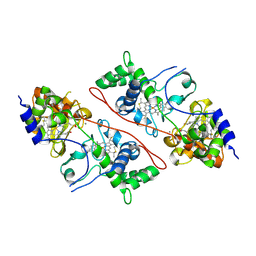 | |
1IVT
 
 | | NMR structures of the C-terminal globular domain of human lamin A/C | | Descriptor: | Lamin A/C | | Authors: | Krimm, I, Ostlund, C, Gilquin, B, Couprie, J, Hossenlopp, P, Mornon, J.P, Bonn, G, Courvalin, J.C, Worman, H.J, Zinn-Justin, S. | | Deposit date: | 2002-03-29 | | Release date: | 2002-08-21 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The Ig-like structure of the C-terminal domain of lamin A/C, mutated in muscular dystrophies, cardiomyopathy, and partial lipodystrophy.
Structure, 10, 2002
|
|
2SEC
 
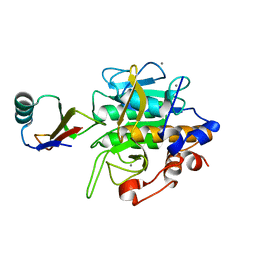 | |
1ELQ
 
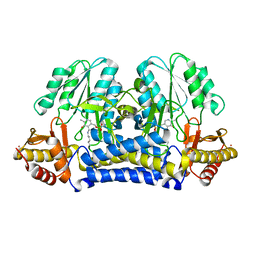 | | CRYSTAL STRUCTURE OF THE CYSTINE C-S LYASE C-DES | | Descriptor: | L-CYSTEINE/L-CYSTINE C-S LYASE, POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Clausen, T, Kaiser, J.T, Steegborn, C, Huber, R, Kessler, D. | | Deposit date: | 2000-03-14 | | Release date: | 2000-04-19 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the cystine C-S lyase from Synechocystis: stabilization of cysteine persulfide for FeS cluster biosynthesis.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1DY9
 
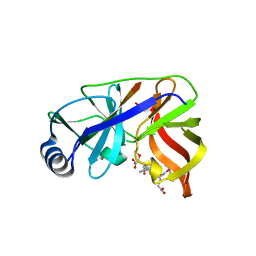 | | Inhibition of the Hepatitis C Virus NS3/4A Protease. The Crystal Structures of Two Protease-Inhibitor Complexes (inhibitor I) | | Descriptor: | N-(tert-butoxycarbonyl)-L-alpha-glutamyl-N-[(1R)-1-(carboxycarbonyl)-3,3-difluoropropyl]-L-leucinamide, NONSTRUCTURAL PROTEIN NS4A (P4), PROTEASE/HELICASE NS3 (P70), ... | | Authors: | Di Marco, S, Rizzi, M, Volpari, C, Walsh, M, Narjes, F, Colarusso, S, De Francesco, R, Matassa, V.G, Sollazzo, M. | | Deposit date: | 2000-01-31 | | Release date: | 2001-01-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition of the Hepatitis C Virus Ns3/4A Protease the Crystal Structures of Two Protease-Inhibitor Complexes
J.Biol.Chem., 275, 2000
|
|
1DXP
 
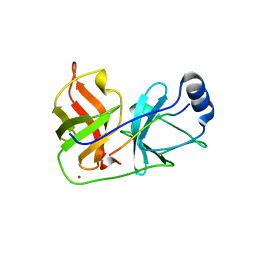 | | Inhibition of the Hepatitis C Virus NS3/4A Protease. The Crystal Structures of Two Protease-Inhibitor Complexes (apo structure) | | Descriptor: | GLYCEROL, NONSTRUCTURAL PROTEIN NS4A (P4), PROTEASE/HELICASE NS3 (P70), ... | | Authors: | Di Marco, S, Rizzi, M, Volpari, C, Walsh, M, Narjes, F, Colarusso, S, De Francesco, R, Matassa, V.G, Sollazzo, M. | | Deposit date: | 2000-01-13 | | Release date: | 2001-01-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibition of the Hepatitis C Virus Ns3/4A Protease the Crystal Structures of Two Protease-Inhibitor Complexes
J.Biol.Chem., 275, 2000
|
|
1EW3
 
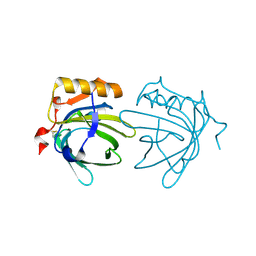 | | CRYSTAL STRUCTURE OF THE MAJOR HORSE ALLERGEN EQU C 1 | | Descriptor: | ALLERGEN EQU C 1 | | Authors: | Lascombe, M.B, Gregoire, C, Poncet, P, Tavares, G.A, Rosinski-Chupin, I, Rabillon, J, Goubran-Botros, H, Mazie, J.C, David, B, Alzari, P.M. | | Deposit date: | 2000-04-21 | | Release date: | 2000-05-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the allergen Equ c 1. A dimeric lipocalin with restricted IgE-reactive epitopes.
J.Biol.Chem., 275, 2000
|
|
3FYE
 
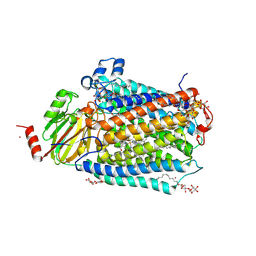 | | Catalytic core subunits (I and II) of cytochrome c oxidase from Rhodobacter sphaeroides in the reduced state | | Descriptor: | CADMIUM ION, CALCIUM ION, COPPER (I) ION, ... | | Authors: | Qin, L, Mills, D.A, Proshlyakov, D.A, Hiser, C, Ferguson-Miller, S. | | Deposit date: | 2009-01-22 | | Release date: | 2009-06-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Redox dependent conformational changes in cytochrome c oxidase suggest a gating mechanism for proton uptake.
Biochemistry, 48, 2009
|
|
1HV2
 
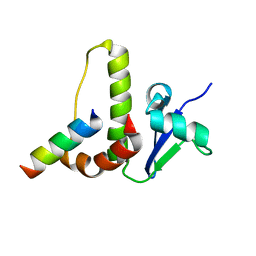 | | SOLUTION STRUCTURE OF YEAST ELONGIN C IN COMPLEX WITH A VON HIPPEL-LINDAU PEPTIDE | | Descriptor: | ELONGIN C, VON HIPPEL-LINDAU DISEASE TUMOR SUPPRESSOR | | Authors: | Botuyan, M.V, Mer, G, Yi, G.-S, Koth, C.M, Case, D.A, Edwards, A.M, Chazin, W.J, Arrowsmith, C.H. | | Deposit date: | 2001-01-05 | | Release date: | 2001-09-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of yeast elongin C in complex with a von Hippel-Lindau peptide.
J.Mol.Biol., 312, 2001
|
|
4FOJ
 
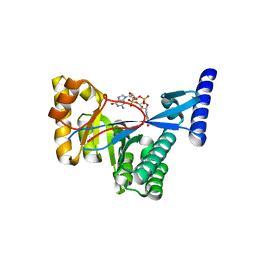 | | 1.55 A Crystal Structure of Xanthomonas citri FimX EAL domain in complex with c-diGMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), FimX | | Authors: | Farah, C.S, Guzzo, C.R. | | Deposit date: | 2012-06-20 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of the PilZ-FimXEAL-c-di-GMP Complex Responsible for the Regulation of Bacterial Type IV Pilus Biogenesis.
J.Mol.Biol., 425, 2013
|
|
4FOU
 
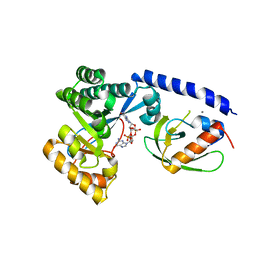 | | Structure of the PilZ-FimX(EAL domain)-c-di-GMP complex responsible for the regulation of bacterial Type IV pilus biogenesis | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, FimX, ... | | Authors: | Farah, C.S, Guzzo, C.R. | | Deposit date: | 2012-06-21 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the PilZ-FimXEAL-c-di-GMP Complex Responsible for the Regulation of Bacterial Type IV Pilus Biogenesis.
J.Mol.Biol., 425, 2013
|
|
1O0E
 
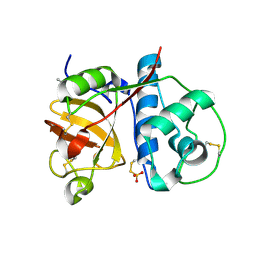 | | 1.9 Angstrom Crystal Structure of a plant cysteine protease Ervatamin C | | Descriptor: | Ervatamin C, THIOSULFATE | | Authors: | Thakurta, P.G, Chakrabarti, C, Biswas, S, Dattagupta, J.K. | | Deposit date: | 2003-02-21 | | Release date: | 2004-03-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of the Unusual Stability and Substrate Specificity of Ervatamin C, a Plant Cysteine Protease from Ervatamia coronaria
Biochemistry, 43, 2004
|
|
3MUR
 
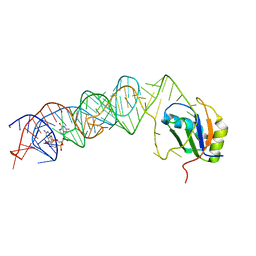 | | Crystal Structure of the C92U mutant c-di-GMP riboswith bound to c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), C92U mutant c-di-GMP riboswitch, MAGNESIUM ION, ... | | Authors: | Strobel, S.A, Smith, K.D. | | Deposit date: | 2010-05-03 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and biochemical determinants of ligand binding by the c-di-GMP riboswitch .
Biochemistry, 49, 2010
|
|
