2VN4
 
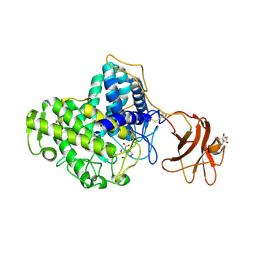 | | Glycoside Hydrolase Family 15 Glucoamylase from Hypocrea jecorina | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bott, R, Sandgren, M, Hansson, H. | | Deposit date: | 2008-01-30 | | Release date: | 2008-05-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Three-Dimensional Structure of an Intact Glycoside Hydrolase Family 15 Glucoamylase from Hypocrea Jecorina.
Biochemistry, 47, 2008
|
|
8BC6
 
 | |
4H7U
 
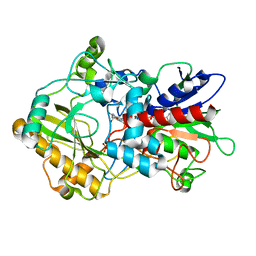 | | Crystal structure of pyranose dehydrogenase from Agaricus meleagris, wildtype | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATE ION, ... | | Authors: | Tan, T.C, Spadiut, O, Divne, C. | | Deposit date: | 2012-09-20 | | Release date: | 2013-08-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1.6 A crystal structure of pyranose dehydrogenase from Agaricus meleagris rationalizes substrate specificity and reveals a flavin intermediate.
Plos One, 8, 2013
|
|
8OQT
 
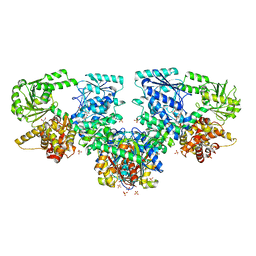 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with Fragment-M-91 | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, 4-bromanylbenzenesulfonic acid, GLYCEROL, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2023-04-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Crystallographic fragment-binding studies of the Mycobacterium tuberculosis trifunctional enzyme suggest binding pockets for the tails of the acyl-CoA substrates at its active sites and a potential substrate-channeling path between them.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8OQS
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with Fragment-M-83 | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, 4-phenylbenzenesulfonic acid, GLYCEROL, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2023-04-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystallographic fragment-binding studies of the Mycobacterium tuberculosis trifunctional enzyme suggest binding pockets for the tails of the acyl-CoA substrates at its active sites and a potential substrate-channeling path between them.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8OQV
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with Fragment-M-109 | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, 4-nitrobenzenesulfonic acid, GLYCEROL, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2023-04-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystallographic fragment-binding studies of the Mycobacterium tuberculosis trifunctional enzyme suggest binding pockets for the tails of the acyl-CoA substrates at its active sites and a potential substrate-channeling path between them.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8OQR
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with Fragment-M-80 | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, 4-cyanobenzenesulfonic acid, GLYCEROL, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2023-04-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic fragment-binding studies of the Mycobacterium tuberculosis trifunctional enzyme suggest binding pockets for the tails of the acyl-CoA substrates at its active sites and a potential substrate-channeling path between them.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8OQL
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with Fragment-M-1 | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, GLYCEROL, Hexafluorophosphate anion, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2023-04-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic fragment-binding studies of the Mycobacterium tuberculosis trifunctional enzyme suggest binding pockets for the tails of the acyl-CoA substrates at its active sites and a potential substrate-channeling path between them.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8OQQ
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with Fragment-M-79 | | Descriptor: | 2-fluoranyl-5-sulfo-benzoic acid, 3-hydroxyacyl-CoA dehydrogenase, GLYCEROL, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2023-04-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystallographic fragment-binding studies of the Mycobacterium tuberculosis trifunctional enzyme suggest binding pockets for the tails of the acyl-CoA substrates at its active sites and a potential substrate-channeling path between them.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8OQU
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with Fragment-M-92 | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, 4-chloranylbenzenesulfonic acid, GLYCEROL, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2023-04-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Crystallographic fragment-binding studies of the Mycobacterium tuberculosis trifunctional enzyme suggest binding pockets for the tails of the acyl-CoA substrates at its active sites and a potential substrate-channeling path between them.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
4J3W
 
 | | Crystal structure of barley limit dextrinase (E510A mutant) in complex with a branched maltohexasaccharide | | Descriptor: | CALCIUM ION, IODIDE ION, Limit dextrinase, ... | | Authors: | Sim, L, Windahl, M.S, Moeller, M.S, Henriksen, A. | | Deposit date: | 2013-02-06 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Oligosaccharide and substrate binding in the starch debranching enzyme barley limit dextrinase
J.Mol.Biol., 427, 2015
|
|
8ORC
 
 | | Mus Musculus Acetylcholinesterase in complex with AL237 | | Descriptor: | 1-[2-(dimethylamino)ethyl]-3-(2-methoxyphenyl)thiourea, 2,5,8,11,14,17,20,23-OCTAOXAPENTACOSAN-25-OL, 2-(2-METHOXYETHOXY)ETHANOL, ... | | Authors: | Ekstrom, F.E, Linusson, A. | | Deposit date: | 2023-04-13 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enzyme Dynamics Determine the Potency and Selectivity of Inhibitors Targeting Disease-Transmitting Mosquitoes.
Acs Infect Dis., 10, 2024
|
|
8ORN
 
 | |
7KF4
 
 | | Crystal structure from SARS-CoV-2 NendoU NSP15 | | Descriptor: | CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Nakamura, A.M, Pereira, H.M, Noske, G.D, Gawriljuk, V.O, Fernandes, R.S, Oliveira, K.I.Z, Oliva, G. | | Deposit date: | 2020-10-13 | | Release date: | 2020-12-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
8OQM
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with Fragment-M-10 | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, 6-[(6-azanyl-4-oxidanyl-naphthalen-2-yl)sulfonylamino]-4-oxidanyl-naphthalene-2-sulfonic acid, 6-azanyl-4-oxidanyl-naphthalene-2-sulfonic acid, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2023-04-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystallographic fragment-binding studies of the Mycobacterium tuberculosis trifunctional enzyme suggest binding pockets for the tails of the acyl-CoA substrates at its active sites and a potential substrate-channeling path between them.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8P4Z
 
 | | Crystal structure of the human CDK7 kinase domain in complex with LDC4297 | | Descriptor: | 2-[(3R)-piperidin-3-yl]oxy-8-propan-2-yl-N-[(2-pyrazol-1-ylphenyl)methyl]pyrazolo[1,5-a][1,3,5]triazin-4-amine, Cyclin-dependent kinase 7, GLYCEROL, ... | | Authors: | Laursen, M, Caing-Carlsson, R, Houssari, R, Javadi, A, Kimbung, Y.R, Murina, V, Orozco-Rodriguez, J.M, Svensson, A, Welin, M, Logan, D, Svensson, B, Walse, B. | | Deposit date: | 2023-05-23 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of the human CDK7 kinase domain in complex with LDC4297
To Be Published
|
|
8PDT
 
 | |
4J3T
 
 | | Crystal structure of barley Limit dextrinase co-crystallized with 25mM maltotetraose | | Descriptor: | CALCIUM ION, CHLORIDE ION, IODIDE ION, ... | | Authors: | Sim, L, Windahl, M.S, Moeller, M.S, Henriksen, A. | | Deposit date: | 2013-02-06 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Oligosaccharide and substrate binding in the starch debranching enzyme barley limit dextrinase
J.Mol.Biol., 427, 2015
|
|
4J3U
 
 | | Crystal structure of barley limit dextrinase in complex with maltosyl-S-betacyclodextrin | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cyclic alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-[alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-1)]6-thio-alpha-D-glucopyranose, ... | | Authors: | Sim, L, Windahl, M.S, Moeller, M.S, Henriksen, A. | | Deposit date: | 2013-02-06 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Oligosaccharide and substrate binding in the starch debranching enzyme barley limit dextrinase
J.Mol.Biol., 427, 2015
|
|
4J3X
 
 | | Crystal structure of barley limit dextrinase (E510A mutant) in complex with a branched maltoheptasaccharide | | Descriptor: | CALCIUM ION, CHLORIDE ION, IODIDE ION, ... | | Authors: | Sim, L, Windahl, M.S, Moeller, M.S, Henriksen, A. | | Deposit date: | 2013-02-06 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Oligosaccharide and substrate binding in the starch debranching enzyme barley limit dextrinase
J.Mol.Biol., 427, 2015
|
|
4KA7
 
 | | Structure of Organellar OligoPeptidase (E572Q) in complex with an endogenous substrate | | Descriptor: | CHLORIDE ION, GLYCEROL, Oligopeptidase A, ... | | Authors: | Berntsson, R.P.-A, Kmiec, B, Teixeira, P.F, Svensson, L.M, Bakali, A, Glaser, E, Stenmark, P. | | Deposit date: | 2013-04-22 | | Release date: | 2013-09-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Organellar oligopeptidase (OOP) provides a complementary pathway for targeting peptide degradation in mitochondria and chloroplasts.
Proc. Natl. Acad. Sci. U.S.A., 110, 2013
|
|
1IZ9
 
 | | Crystal Structure of Malate Dehydrogenase from Thermus thermophilus HB8 | | Descriptor: | MALATE DEHYDROGENASE | | Authors: | Hirose, R, Hasegawa, T, Yamano, A, Kuramitsu, S, Hamada, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-10-01 | | Release date: | 2002-10-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Malate Dehydrogenase from Thermus themrophilus HB8
To be published
|
|
7NEO
 
 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 15 | | Descriptor: | 2-cyclobutyl-7-(5-fluoropyridin-3-yl)-5,7-diazaspiro[3.4]octane-6,8-dione, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Talibov, V.O. | | Deposit date: | 2021-02-04 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
7NBT
 
 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 21 | | Descriptor: | 2-(benzotriazol-1-yl)-1-[(4~{S})-4-methyl-6,7-dihydro-4~{H}-thieno[3,2-c]pyridin-5-yl]ethanone, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Talibov, V.O. | | Deposit date: | 2021-01-27 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
7NCZ
 
 | | Crystal structure of Paradendryphiella salina PL7A alginate lyase mutant Y223F in complex with hexa-mannuronic acid | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alginate lyase (PL7), beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Fredslund, F, Welner, D.H, Wilkens, C. | | Deposit date: | 2021-01-29 | | Release date: | 2022-03-02 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Unraveling the molecular mechanism of polysaccharide lyases for efficient alginate degradation.
Nat Commun, 16, 2025
|
|
