1U1I
 
 | | Myo-inositol phosphate synthase mIPS from A. fulgidus | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Stieglitz, K.A, Yang, H, Roberts, M.F, Stec, B. | | Deposit date: | 2004-07-15 | | Release date: | 2004-08-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reaching for Mechanistic Consensus Across Life Kingdoms: Structure and Insights into Catalysis of the myo-Inositol-1-phosphate Synthase (mIPS) from Archaeoglobus fulgidus
Biochemistry, 44, 2005
|
|
2ZGX
 
 | | Thrombin Inhibition | | Descriptor: | 1-[(2R)-2-aminobutanoyl]-N-(4-carbamimidoylbenzyl)-L-prolinamide, Hirudin variant-1, SODIUM ION, ... | | Authors: | Baum, B, Heine, A, Klebe, G. | | Deposit date: | 2008-01-28 | | Release date: | 2008-12-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Think twice: understanding the high potency of bis(phenyl)methane inhibitors of thrombin
J.Mol.Biol., 391, 2009
|
|
4NR7
 
 | | Crystal structure of the bromodomain of human CREBBP in complex with an isoxazolyl-benzimidazole ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(3-chloro-4-methoxyphenyl)ethyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(2S)-2-(morpholin-4-yl)propyl]-1H-benzimidazole, CREB-binding protein | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Hay, D, Fedorov, O, Martin, S, Krojer, T, Nowak, R, von Delft, F, Brennan, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-11-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of the bromodomain of human CREBBP in complex with an isoxazolyl-benzimidazole ligand
To be Published
|
|
4NYF
 
 | | HIV integrase in complex with inhibitor | | Descriptor: | (2S)-tert-butoxy[4-(4-chlorophenyl)-2-methylquinolin-3-yl]ethanoic acid, CADMIUM ION, Integrase | | Authors: | Coulombe, R, Fader, L. | | Deposit date: | 2013-12-10 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of BI 224436, a Noncatalytic Site Integrase Inhibitor (NCINI) of HIV-1.
ACS Med Chem Lett, 5, 2014
|
|
4F3V
 
 | |
5TLE
 
 | |
4BV3
 
 | | CRYSTAL STRUCTURE OF SIRT3 IN COMPLEX WITH THE INHIBITOR EX-527 AND NAD | | Descriptor: | (1S)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1- carboxamide, ADENOSINE-5-DIPHOSPHORIBOSE, CHLORIDE ION, ... | | Authors: | Gertz, M, Nguyen, N.T.T, Weyand, M, Steegborn, C. | | Deposit date: | 2013-06-24 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ex-527 Inhibits Sirtuins by Exploiting Their Unique Nad+-Dependent Deacetylation Mechanism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3Q6G
 
 | | Crystal structure of Fab of rhesus mAb 2.5B specific for quaternary neutralizing epitope of HIV-1 gp120 | | Descriptor: | Heavy chain of Fab of rhesus mAb 2.5B, Light chain of Fab of rhesus mAb 2.5B | | Authors: | Spurrier, B, Sampson, J, Totrov, M, Li, H, O'Neal, T, William, C, Robinson, J, Gorny, M.K, Zolla-Pazner, S, Kong, X.P. | | Deposit date: | 2010-12-31 | | Release date: | 2011-05-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structural Analysis of Human and Macaque mAbs 2909 and 2.5B: Implications for the Configuration of the Quaternary Neutralizing Epitope of HIV-1 gp120.
Structure, 19, 2011
|
|
1U1E
 
 | | Structure of e. coli uridine phosphorylase complexed to 5(phenylseleno)acyclouridine (PSAU) | | Descriptor: | 1-((2-HYDROXYETHOXY)METHYL)-5-(PHENYLSELANYL)PYRIMIDINE-2,4(1H,3H)-DIONE, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Bu, W, Settembre, E.C, Ealick, S.E. | | Deposit date: | 2004-07-15 | | Release date: | 2005-07-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural basis for inhibition of Escherichia coli uridine phosphorylase by 5-substituted acyclouridines.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4J59
 
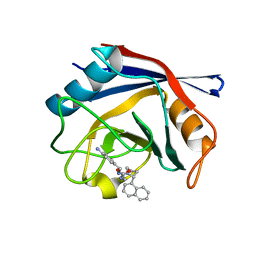 | | Human Cyclophilin D Complexed with an Inhibitor | | Descriptor: | 1-(4-aminobenzyl)-3-{2-[(2R)-2-(naphthalen-1-yl)pyrrolidin-1-yl]-2-oxoethyl}urea, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Gelin, M, Colliandre, L, Bessin, Y, Guichou, J.F. | | Deposit date: | 2013-02-08 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Fragment-based discovery of a new family of non-peptidic small-molecule cyclophilin inhibitors with potent antiviral activities.
Nat Commun, 7, 2016
|
|
4J67
 
 | | Crystal structure of Ribonuclease A soaked in 50% 1,6-Hexanediol: One of twelve in MSCS set | | Descriptor: | HEXANE-1,6-DIOL, Ribonuclease pancreatic, SULFATE ION | | Authors: | Kearney, B.M, Dechene, M, Swartz, P.D, Mattos, C. | | Deposit date: | 2013-02-11 | | Release date: | 2014-01-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | DRoP: A program for analysis of water structure on protein surfaces
to be published
|
|
2WOQ
 
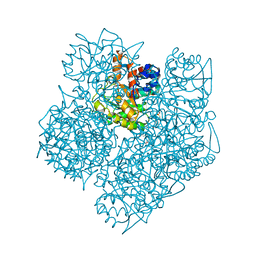 | | Porphobilinogen Synthase (HemB) in Complex with 5-acetamido-4- oxohexanoic acid (Alaremycin 2) | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ALAREMYCIN 2, ... | | Authors: | Heinemann, I.U, Schulz, C, Schubert, W.-D, Heinz, D.W, Wang, Y.-G, Kobayashi, Y, Awa, Y, Wachi, M, Jahn, D, Jahn, M. | | Deposit date: | 2009-07-27 | | Release date: | 2009-10-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the heme biosynthetic Pseudomonas aeruginosa porphobilinogen synthase in complex with the antibiotic alaremycin.
Antimicrob. Agents Chemother., 54, 2010
|
|
2WQK
 
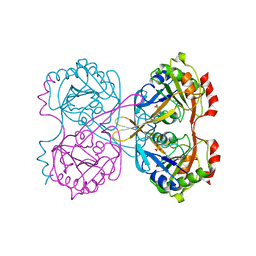 | | Crystal Structure of Sure Protein from Aquifex aeolicus | | Descriptor: | 5'-NUCLEOTIDASE SURE, SODIUM ION, SULFATE ION | | Authors: | Antonyuk, S.V, Ellis, M.J, Strange, R.W, Hasnain, S.S, Bessho, Y, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-08-23 | | Release date: | 2009-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Sure Protein from Aquifex Aeolicus Vf5 at 1.5 A Resolution.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
2ZAZ
 
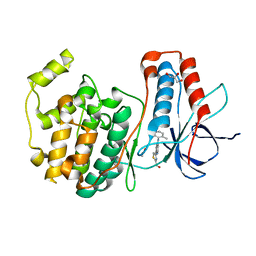 | | Crystal structure of P38 in complex with 4-anilino quinoline inhibitor | | Descriptor: | 4-{4-[(5-hydroxy-2-methylphenyl)amino]quinolin-7-yl}-1,3-thiazole-2-carbaldehyde, ACETATE ION, GLYCEROL, ... | | Authors: | Somers, D.O. | | Deposit date: | 2007-10-12 | | Release date: | 2008-01-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biphenyl amide p38 kinase inhibitors 1: Discovery and binding mode
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2ZSE
 
 | | Pantothenate kinase from Mycobacterium tuberculosis (MtPanK) in complex with AMPPCP and Pantothenate | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, GLYCEROL, ... | | Authors: | Chetnani, B, Das, S, Kumar, P, Surolia, A, Vijayan, M. | | Deposit date: | 2008-09-05 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mycobacterium tuberculosis pantothenate kinase: possible changes in location of ligands during enzyme action
Acta Crystallogr.,Sect.D, 65, 2009
|
|
6K4X
 
 | | Crystal structure of SMB-1 metallo-beta-lactamase in a complex with ASB | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-azanyl-2-sulfanyl-benzoic acid, Metallo-beta-lactamase, ... | | Authors: | Wachino, J. | | Deposit date: | 2019-05-27 | | Release date: | 2019-08-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | 4-Amino-2-Sulfanylbenzoic Acid as a Potent Subclass B3 Metallo-beta-Lactamase-Specific Inhibitor Applicable for Distinguishing Metallo-beta-Lactamase Subclasses.
Antimicrob.Agents Chemother., 63, 2019
|
|
7WWC
 
 | | Crystal structure of beta-1,3(4)-glucanase with Laminaritriose | | Descriptor: | beta-1,3(4)-glucanase, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Jiang, Z.Q, Ma, J.W. | | Deposit date: | 2022-02-12 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2021625 Å) | | Cite: | Crystal structure of beta-1,3(4)-glucanase with Laminaritriose
To Be Published
|
|
1TOP
 
 | |
1MC3
 
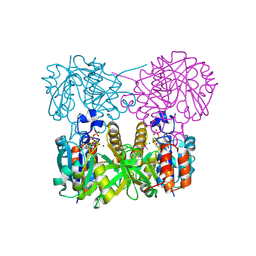 | | CRYSTAL STRUCTURE OF RFFH | | Descriptor: | GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, MAGNESIUM ION, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Sivaraman, J, Sauve, V, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2002-08-05 | | Release date: | 2002-11-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Escherichia coli Glucose-1-Phosphate Thymidylyltransferase (RffH) Complexed with dTTP and Mg2+
J.BIOL.CHEM., 277, 2002
|
|
3UX4
 
 | |
1UBQ
 
 | |
2ZFP
 
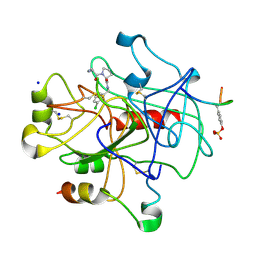 | | Thrombin Inibition | | Descriptor: | 1-[(2R)-2-aminobutanoyl]-N-(3-chlorobenzyl)-L-prolinamide, Hirudin variant-1, SODIUM ION, ... | | Authors: | Baum, B, Heine, A, Klebe, G. | | Deposit date: | 2008-01-08 | | Release date: | 2008-12-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Think twice: understanding the high potency of bis(phenyl)methane inhibitors of thrombin
J.Mol.Biol., 391, 2009
|
|
2ONZ
 
 | | Structure of K57A hPNMT with inhibitor 7-(N-4-chlorophenylaminosulfonyl)-THIQ and AdoHcy | | Descriptor: | N-(4-CHLOROPHENYL)-1,2,3,4-TETRAHYDROISOQUINOLINE-7-SULFONAMIDE, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2007-01-25 | | Release date: | 2007-10-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Enzyme Adaptation to Inhibitor Binding: A Cryptic Binding Site in Phenylethanolamine N-Methyltransferase
J.Med.Chem., 50, 2007
|
|
4OV2
 
 | |
4C9S
 
 | | BACTERIAL CHALCONE ISOMERASE IN open CONFORMATION FROM EUBACTERIUM RAMULUS AT 1.8 A RESOLUTION | | Descriptor: | CHALCONE ISOMERASE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Thomsen, M, Palm, G.J, Hinrichs, W. | | Deposit date: | 2013-10-03 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Catalytic Mechanism of the Evolutionarily Unique Bacterial Chalcone Isomerase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
