3VFU
 
 | | crystal structure of HLA B*3508 LPEP-P7Ala, peptide mutant P7-ala | | Descriptor: | Beta-2-microglobulin, LPEP peptide from EBV, P7A, ... | | Authors: | Liu, Y.C, Rossjohn, J, Gras, S. | | Deposit date: | 2012-01-10 | | Release date: | 2012-02-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Energetic Basis Underpinning T-cell Receptor Recognition of a Super-bulged Peptide Bound to a Major Histocompatibility Complex Class I Molecule.
J.Biol.Chem., 287, 2012
|
|
3VG5
 
 | | Barium derivative of human LFABP | | Descriptor: | BARIUM ION, Fatty acid-binding protein, liver, ... | | Authors: | Sharma, A, Yogavel, M, Sharma, A. | | Deposit date: | 2011-08-03 | | Release date: | 2012-06-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Utility of anion and cation combinations for phasing of protein structures.
J.Struct.Funct.Genom., 13, 2012
|
|
3JY6
 
 | | Crystal structure of LacI Transcriptional regulator from Lactobacillus brevis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Transcriptional regulator, ... | | Authors: | Syed Ibrahim, B, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-21 | | Release date: | 2009-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of LacI Transcriptional regulator from Lactobacillus brevis
To be Published
|
|
3VMK
 
 | | 3-isopropylmalate dehydrogenase from Shewanella benthica DB21 MT-2 | | Descriptor: | 3-ISOPROPYLMALIC ACID, 3-isopropylmalate dehydrogenase, CHLORIDE ION, ... | | Authors: | Nagae, T, Watanabe, N. | | Deposit date: | 2011-12-13 | | Release date: | 2012-02-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural analysis of 3-isopropylmalate dehydrogenase from the obligate piezophile Shewanella benthica DB21MT-2 and the nonpiezophile Shewanella oneidensis MR-1
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3JYU
 
 | | Crystal structure of the N-terminal domains of the ubiquitin specific peptidase 4 (USP4) | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, Ubiquitin carboxyl-terminal hydrolase | | Authors: | Bacik, J.P, Avvakumov, G, Walker, J.R, Xue, S, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-22 | | Release date: | 2009-10-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of the N-terminal domains of the ubiquitin specific peptidase 4 (USP4)
To be Published
|
|
1WAE
 
 | | Crystal structure of H129V Mutant of Alcaligenes Xylosoxidans Nitrite Reductase | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, ZINC ION | | Authors: | Ellis, M.J, Antonyuk, S.V, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2004-10-26 | | Release date: | 2004-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Observation of an Unprecedented Cu Bis-His Site: Crystal Structure of the H129V Mutant of Nitrite Reductase
Inorg.Chem., 43, 2004
|
|
3VNF
 
 | | Structure of the ebolavirus protein VP24 from Sudan | | Descriptor: | Membrane-associated protein VP24 | | Authors: | Zhang, A.P.P. | | Deposit date: | 2012-01-12 | | Release date: | 2012-04-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The ebola virus interferon antagonist VP24 directly binds STAT1 and has a novel, pyramidal fold
Plos Pathog., 8, 2012
|
|
3JTZ
 
 | | Structure of the arm-type binding domain of HPI integrase | | Descriptor: | Integrase, SODIUM ION | | Authors: | Szwagierczak, A, Antonenka, U, Popowicz, G.M, Sitar, T, Holak, T.A, Rakin, A. | | Deposit date: | 2009-09-14 | | Release date: | 2009-10-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of the arm-type binding domains of HPI and HAI7 integrases
J.Biol.Chem., 284, 2009
|
|
3VOD
 
 | | Crystal Structure of mutant MarR C80S from E.coli | | Descriptor: | Multiple antibiotic resistance protein marR | | Authors: | Lou, H, Zhu, R, Hao, Z. | | Deposit date: | 2012-01-21 | | Release date: | 2013-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The multiple antibiotic resistance regulator MarR is a copper sensor in Escherichia coli.
Nat.Chem.Biol., 10, 2014
|
|
3VOR
 
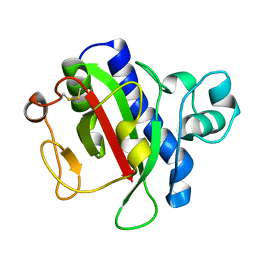 | | Crystal Structure Analysis of the CofA | | Descriptor: | CFA/III pilin | | Authors: | Fukakusa, S, Kawahara, K, Nakamura, S, Iwasita, T, Baba, S, Nishimura, M, Kobayashi, Y, Honda, T, Iida, T, Taniguchi, T, Ohkubo, T. | | Deposit date: | 2012-02-06 | | Release date: | 2012-09-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Structure of the CFA/III major pilin subunit CofA from human enterotoxigenic Escherichia coli determined at 0.90 A resolution by sulfur-SAD phasing
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3VHC
 
 | | Hsp90 alpha N-terminal domain in complex with a macrocyclic inhibitor | | Descriptor: | 4-amino-20,22-dimethyl-13-oxa-7-thia-3,5,17-triazatetracyclo[17.3.1.1~2,6~.1~8,12~]pentacosa-1(23),2(25),3,5,8(24),9,11,19,21-nonaen-18-one, Heat shock protein HSP 90-alpha, MAGNESIUM ION | | Authors: | Fukami, T.A, Ono, N. | | Deposit date: | 2011-08-24 | | Release date: | 2012-07-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Design and synthesis of novel macrocyclic 2-amino-6-arylpyrimidine Hsp90 inhibitors
Bioorg.Med.Chem.Lett., 22, 2012
|
|
1W3X
 
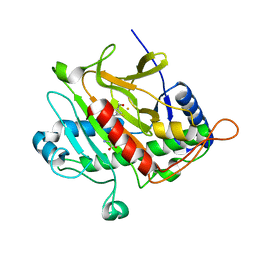 | | Isopenicillin N synthase d-(L-a-aminoadipoyl)-(3R)-methyl-L-cysteine D-a-hydroxyisovaleryl ester complex (Oxygen exposed 5 minutes 20 bar) | | Descriptor: | FE (II) ION, ISOPENICILLIN N SYNTHETASE, N~6~-[(1R)-1-({[(1R,2R)-1-CARBOXY-3-HYDROXY-2-METHYLPROPYL]OXY}CARBONYL)-2-MERCAPTOPROP-2-EN-1-YL]-6-OXO-L-LYSINE | | Authors: | Daruzzaman, A, Clifton, I.J, Rutledge, P.J. | | Deposit date: | 2004-07-20 | | Release date: | 2005-12-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Unexpected Oxidation of a Depsipeptide Substrate Analogue in Crystalline Isopenicillin N Synthase.
Chembiochem, 7, 2006
|
|
1W6W
 
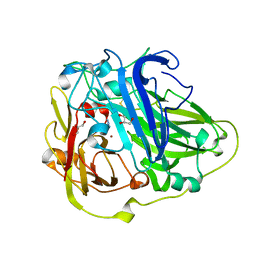 | | 3D structure of CotA incubated with sodium azide | | Descriptor: | AZIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Bento, I, Martins, L.O, Lopes, G.G, Carrondo, M.A, Lindley, P.F. | | Deposit date: | 2004-08-24 | | Release date: | 2005-10-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dioxygen Reduction by Multi-Copper Oxidases; a Structural Perspective.
Dalton Trans., 7, 2005
|
|
3VQS
 
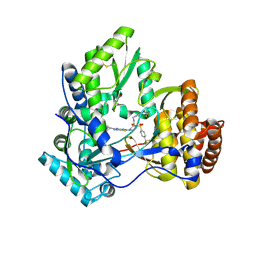 | | Crystal structure of HCV NS5B RNA polymerase with a novel piperazine inhibitor | | Descriptor: | (2R)-4-(5-cyclopropyl[1,3]thiazolo[4,5-d]pyrimidin-2-yl)-N-[3-fluoro-4-(trifluoromethoxy)benzyl]-1-{[4-(trifluoromethyl)phenyl]sulfonyl}piperazine-2-carboxamide, CHLORIDE ION, RNA-directed RNA polymerase | | Authors: | Adachi, T, Doi, S, Ando, I, Sugimoto, K, Orita, T, Nomura, A, Kamada, M. | | Deposit date: | 2012-03-30 | | Release date: | 2012-08-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Preclinical Characterization of JTK-853, a Novel Nonnucleoside Inhibitor of the Hepatitis C Virus RNA-Dependent RNA Polymerase.
Antimicrob.Agents Chemother., 56, 2012
|
|
3VKP
 
 | | Assimilatory nitrite reductase (Nii3) - NO2 complex from tobbaco leaf analysed with low X-ray dose | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, NITRITE ION, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-11-20 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The reductive reaction mechanism of tobacco nitrite reductase derived from a combination of crystal structures and ultraviolet-visible microspectroscopy
Proteins, 80, 2012
|
|
3VL7
 
 | |
1W1U
 
 | |
3VRQ
 
 | | Crystal structure of the tyrosine kinase binding domain of Cbl-c (PL mutant) | | Descriptor: | CALCIUM ION, Signal transduction protein CBL-C | | Authors: | Takeshita, K, Tezuka, T, Isozaki, Y, Yamashita, E, Suzuki, M, Yamanashi, Y, Yamamoto, T, Nakagawa, A. | | Deposit date: | 2012-04-13 | | Release date: | 2013-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural flexibility regulates phosphopeptide-binding activity of the tyrosine kinase binding domain of Cbl-c.
J.Biochem., 152, 2012
|
|
3VSF
 
 | | Crystal structure of 1,3Gal43A, an exo-beta-1,3-Galactanase from Clostridium thermocellum | | Descriptor: | GLYCEROL, Ricin B lectin | | Authors: | Jiang, D, Fan, J, Wang, X, Zhao, Y, Huang, B, Zhang, X.C. | | Deposit date: | 2012-04-25 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.757 Å) | | Cite: | Crystal structure of 1,3Gal43A, an exo-beta-1,3-galactanase from Clostridium thermocellum
J.Struct.Biol., 180, 2012
|
|
3VTF
 
 | | Structure of a UDP-glucose dehydrogenase from the hyperthermophilic archaeon Pyrobaculum islandicum | | Descriptor: | 1,2-ETHANEDIOL, UDP-glucose 6-dehydrogenase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Sakuraba, H, Ohshima, T, Yoneda, K. | | Deposit date: | 2012-05-29 | | Release date: | 2012-09-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a UDP-glucose dehydrogenase from the hyperthermophilic archaeon Pyrobaculum islandicum.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
2PI8
 
 | | Crystal structure of E. coli MltA with bound chitohexaose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Membrane-bound lytic murein transglycosylase A, PHOSPHATE ION | | Authors: | van Straaten, K.E, Barends, T.R.M, Dijkstra, B.W, Thunnissen, A.M.W.H. | | Deposit date: | 2007-04-13 | | Release date: | 2007-05-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of Escherichia coli Lytic transglycosylase MltA with bound chitohexaose: implications for peptidoglycan binding and cleavage
J.Biol.Chem., 282, 2007
|
|
3VTM
 
 | | Structure of heme transport protein IsdH-NEAT3 from S. aureus in complex with Indium-porphyrin | | Descriptor: | GLYCEROL, Iron-regulated surface determinant protein H, PROTOPORPHYRIN IX CONTAINING INDIUM | | Authors: | Vu, N.T, Caaveiro, J.M.M, Moriwaki, Y, Tsumoto, K. | | Deposit date: | 2012-05-31 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Selective binding of antimicrobial porphyrins to the heme-receptor IsdH-NEAT3 of Staphylococcus aureus
Protein Sci., 22, 2013
|
|
3VP5
 
 | | X-ray Crystal Structure of Wild Type HrtR in the Holo Form | | Descriptor: | CACODYLATE ION, PROTOPORPHYRIN IX CONTAINING FE, Transcriptional regulator | | Authors: | Sawai, H, Sugimoto, H, Shiro, Y, Aono, S. | | Deposit date: | 2012-02-23 | | Release date: | 2012-07-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Transcriptional Regulation of Heme Homeostasis in Lactococcus lactis.
J.Biol.Chem., 287, 2012
|
|
3VTX
 
 | | Crystal structure of MamA protein | | Descriptor: | GLYCEROL, MamA | | Authors: | Zeytuni, N, Baran, D, Davidov, G, Zarivach, R. | | Deposit date: | 2012-06-08 | | Release date: | 2012-10-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Inter-phylum structural conservation of the magnetosome-associated TPR-containing protein, MamA
J.Struct.Biol., 180, 2012
|
|
3VQT
 
 | | Crystal structure analysis of the translation factor RF3 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Peptide chain release factor 3 | | Authors: | Kihira, K, Shomura, Y, Shibata, N, Kitamura, M, Higuchi, Y. | | Deposit date: | 2012-03-30 | | Release date: | 2012-09-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure analysis of the translation factor RF3 (release factor 3)
Febs Lett., 586, 2012
|
|
