1JLT
 
 | | Vipoxin Complex | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, PHOSPHOLIPASE A2, ... | | Authors: | Banumathi, S, Rajashankar, K.R, Notzel, C, Aleksiev, B, Singh, T.P, Genov, N, Betzel, C. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the neurotoxic complex vipoxin at 1.4 A resolution.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
4PXO
 
 | | Crystal structure of Maleylacetoacetate isomerase from Methylobacteriu extorquens AM1 WITH BOUND MALONATE AND GSH (TARGET EFI-507068) | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, MALONIC ACID, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-03-24 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of glutathione s-transferase zeta from Methylobacterium extorquens (TARGET EFI-507068)
To be Published
|
|
4BDD
 
 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | Descriptor: | 1,2-ETHANEDIOL, 1-(TERT-BUTYL)-3-(QUINOXALIN-6-YL)UREA, NITRATE ION, ... | | Authors: | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
5ETS
 
 | | S. aureus 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase complexed with AMPCPP and inhibitor at 1.95 angstrom resolution | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 2-azanyl-8-[(4-chlorophenyl)methylsulfanyl]-1,9-dihydropurin-6-one, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Dennis, M.L, Peat, T.S, Swarbrick, J.D. | | Deposit date: | 2015-11-18 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for the Selective Binding of Inhibitors to 6-Hydroxymethyl-7,8-dihydropterin Pyrophosphokinase from Staphylococcus aureus and Escherichia coli.
J.Med.Chem., 59, 2016
|
|
5ETT
 
 | | S. aureus 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase complexed with AMPCPP and inhibitor at 1.55 angstrom resolution | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 4-[(2-azanyl-6-oxidanylidene-1,9-dihydropurin-8-yl)sulfanylmethyl]-3-fluoranyl-benzenecarbonitrile, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Dennis, M.L, Peat, T.S, Swarbrick, J.D. | | Deposit date: | 2015-11-18 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Basis for the Selective Binding of Inhibitors to 6-Hydroxymethyl-7,8-dihydropterin Pyrophosphokinase from Staphylococcus aureus and Escherichia coli.
J.Med.Chem., 59, 2016
|
|
5ETP
 
 | | E. coli 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase complexed with AMPCPP and inhibitor at 1.05 angstrom resolution | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 2-azanyl-8-[2-(4-bromophenyl)-2-oxidanylidene-ethyl]sulfanyl-1,9-dihydropurin-6-one, CALCIUM ION, ... | | Authors: | Dennis, M.L, Peat, T.S, Swarbrick, J.D. | | Deposit date: | 2015-11-17 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural Basis for the Selective Binding of Inhibitors to 6-Hydroxymethyl-7,8-dihydropterin Pyrophosphokinase from Staphylococcus aureus and Escherichia coli.
J.Med.Chem., 59, 2016
|
|
1UYK
 
 | | Human Hsp90-alpha with 8-Benzo[1,3]dioxol-,5-ylmethyl-9-butyl-2-fluoro-9H-purin-6-ylamine | | Descriptor: | 8-BENZO[1,3]DIOXOL-,5-YLMETHYL-9-BUTYL-2-FLUORO-9H-PURIN-6-YLAMINE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Wright, L, Barril, X, Dymock, B, Sheridan, L, Surgenor, A, Beswick, M, Drysdale, M, Collier, A, Massey, A, Davies, N, Fink, A, Fromont, C, Aherne, W, Boxall, K, Sharp, S, Workman, P, Hubbard, R.E. | | Deposit date: | 2004-03-02 | | Release date: | 2004-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Activity Relationships in Purine-Based Inhibitor Binding to Hsp90 Isoforms
Chem.Biol., 11, 2004
|
|
5ETR
 
 | | S. aureus 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase complexed with AMPCPP and inhibitor at 1.32 angstrom resolution | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 2-azanyl-8-[(4-fluorophenyl)methylsulfanyl]-1,7-dihydropurin-6-one, CHLORIDE ION, ... | | Authors: | Dennis, M.L, Peat, T.S, Swarbrick, J.D. | | Deposit date: | 2015-11-18 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural Basis for the Selective Binding of Inhibitors to 6-Hydroxymethyl-7,8-dihydropterin Pyrophosphokinase from Staphylococcus aureus and Escherichia coli.
J.Med.Chem., 59, 2016
|
|
4GZV
 
 | |
3BRO
 
 | | Crystal structure of the transcription regulator MarR from Oenococcus oeni PSU-1 | | Descriptor: | CHLORIDE ION, GLYCEROL, Transcriptional regulator | | Authors: | Kim, Y, Volkart, L, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-21 | | Release date: | 2008-01-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal Structure of the Transcription Regulator MarR from Oenococcus oeni PSU-1.
To be Published
|
|
5K6X
 
 | | Sidekick-2 immunoglobulin domains 1-4, crystal form 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Goodman, K.M, Mannepalli, S, Honig, B, Shapiro, L. | | Deposit date: | 2016-05-25 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis of sidekick-mediated cell-cell adhesion and specificity.
Elife, 5, 2016
|
|
4Q9N
 
 | | Crystal structure of Chlamydia trachomatis enoyl-ACP reductase (FabI) in complex with NADH and AFN-1252 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Enoyl-[acyl-carrier-protein] reductase [NADH], N-methyl-N-[(3-methyl-1-benzofuran-2-yl)methyl]-3-(7-oxo-5,6,7,8-tetrahydro-1,8-naphthyridin-3-yl)propanamide | | Authors: | Yao, J, Abdelrahman, Y, Robertson, R.M, Cox, J.V, Belland, R.J, White, S.W, Rock, C.O. | | Deposit date: | 2014-05-01 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | Type II Fatty Acid Synthesis Is Essential for the Replication of Chlamydia trachomatis.
J.Biol.Chem., 289, 2014
|
|
2VLH
 
 | | Quinonoid intermediate of Citrobacter freundii tyrosine phenol-lyase formed with methionine | | Descriptor: | (2E)-2-{[(Z)-{3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4(1H)-YLIDENE}METHYL]IMINO}-4-(METHYLSULFANYL)BUTANOIC ACID, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, POTASSIUM ION, ... | | Authors: | Milic, D, Demidkina, T.V, Matkovic-Calogovic, D, Antson, A.A. | | Deposit date: | 2008-01-14 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Insights Into the Catalytic Mechanism of Tyrosine Phenol-Lyase from X-Ray Structures of Quinonoid Intermediates.
J.Biol.Chem., 283, 2008
|
|
4H8Z
 
 | | Radiation damage study of lysozyme - 0.21 MGy | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C | | Authors: | Sutton, K.A, Snell, E.H. | | Deposit date: | 2012-09-24 | | Release date: | 2013-05-15 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.1998 Å) | | Cite: | Insights into the mechanism of X-ray-induced disulfide-bond cleavage in lysozyme crystals based on EPR, optical absorption and X-ray diffraction studies.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4H9E
 
 | | Radiation damage study of lysozyme - 0.84 MGy | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C | | Authors: | Sutton, K.A, Snell, E.H. | | Deposit date: | 2012-09-24 | | Release date: | 2013-05-15 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.1998 Å) | | Cite: | Insights into the mechanism of X-ray-induced disulfide-bond cleavage in lysozyme crystals based on EPR, optical absorption and X-ray diffraction studies.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4H8Y
 
 | | Radiation damage study of lysozyme- 0.14 MGy | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C | | Authors: | Sutton, K.A, Snell, E.H. | | Deposit date: | 2012-09-24 | | Release date: | 2013-05-15 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.1998 Å) | | Cite: | Insights into the mechanism of X-ray-induced disulfide-bond cleavage in lysozyme crystals based on EPR, optical absorption and X-ray diffraction studies.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4H93
 
 | | Radiation damage study of lysozyme - 0.49 MGy | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C | | Authors: | Sutton, K.A, Snell, E.H. | | Deposit date: | 2012-09-24 | | Release date: | 2013-05-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2003 Å) | | Cite: | Insights into the mechanism of X-ray-induced disulfide-bond cleavage in lysozyme crystals based on EPR, optical absorption and X-ray diffraction studies.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4H9A
 
 | | Radiation damage study of lysozyme - 0.63 MGy | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C | | Authors: | Sutton, K.A, Snell, E.H. | | Deposit date: | 2012-09-24 | | Release date: | 2013-05-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.1997 Å) | | Cite: | Insights into the mechanism of X-ray-induced disulfide-bond cleavage in lysozyme crystals based on EPR, optical absorption and X-ray diffraction studies.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1HRI
 
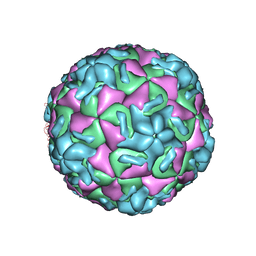 | | STRUCTURE DETERMINATION OF ANTIVIRAL COMPOUND SCH 38057 COMPLEXED WITH HUMAN RHINOVIRUS 14 | | Descriptor: | 1-[6-(2-CHLORO-4-METHYXYPHENOXY)-HEXYL]-IMIDAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Zhang, A, Nanni, R.G, Oren, D.A, Arnold, E. | | Deposit date: | 1992-10-01 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure determination of antiviral compound SCH 38057 complexed with human rhinovirus 14.
J.Mol.Biol., 230, 1993
|
|
7XOC
 
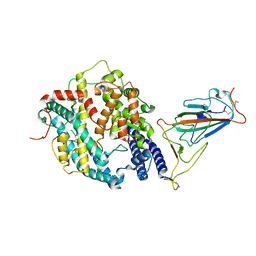 | | SARS-CoV-2 Omicron BA.2 Variant RBD complexed with mouse ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2022-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
7K6N
 
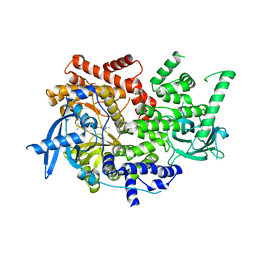 | | Crystal structure of PI3Kalpha selective Inhibitor 11-1575 | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, tert-butyl (3S)-3-[4-(2-aminopyrimidin-5-yl)-2-(morpholin-4-yl)-5,6-dihydro-7H-pyrrolo[2,3-d]pyrimidin-7-yl]-3-methylpyrrolidine-1-carboxylate | | Authors: | Chen, P, Brooun, A, Deng, Y.L, Grodsky, N, Kaiser, S.E. | | Deposit date: | 2020-09-21 | | Release date: | 2021-01-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structure-Based Drug Design and Synthesis of PI3K alpha-Selective Inhibitor (PF-06843195).
J.Med.Chem., 64, 2021
|
|
3C5L
 
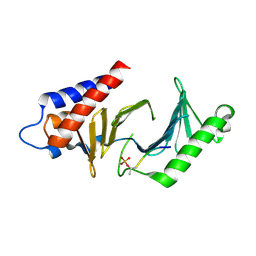 | |
2O73
 
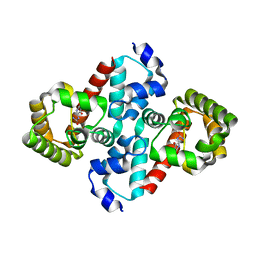 | | Structure of OHCU decarboxylase in complex with allantoin | | Descriptor: | 1-(2,5-DIOXO-2,5-DIHYDRO-1H-IMIDAZOL-4-YL)UREA, OHCU decarboxylase | | Authors: | Cendron, L, Berni, R, Folli, C, Ramazzina, I, Percudani, R, Zanotti, G. | | Deposit date: | 2006-12-10 | | Release date: | 2007-04-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase provides insights into the mechanism of uric acid degradation.
J.Biol.Chem., 282, 2007
|
|
5EW8
 
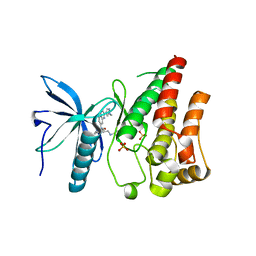 | | FIBROBLAST GROWTH FACTOR RECEPTOR 1 IN COMPLEX WITH JNJ-4275693 | | Descriptor: | Fibroblast growth factor receptor 1, SULFATE ION, ~{N}'-(3,5-dimethoxyphenyl)-~{N}'-[3-(1-methylpyrazol-4-yl)quinoxalin-6-yl]-~{N}-propan-2-yl-ethane-1,2-diamine | | Authors: | Ogg, D, Breed, J. | | Deposit date: | 2015-11-20 | | Release date: | 2016-03-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Landscape of activating cancer mutations in FGFR kinases and their differential responses to inhibitors in clinical use.
Oncotarget, 7, 2016
|
|
4PTN
 
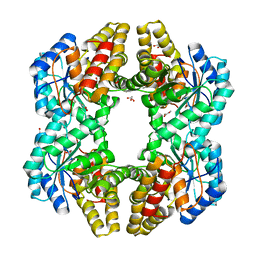 | | Crystal Structure of YagE, a KDG aldolase protein in complex with Magnesium cation coordinated L-glyceraldehyde | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, L-glyceraldehyde, ... | | Authors: | Manoj Kumar, P, Baskar, V, Manicka, S, Krishnaswamy, S. | | Deposit date: | 2014-03-11 | | Release date: | 2014-12-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structure of YagE, a putative DHDPS-like protein from Escherichia coli K12.
Proteins, 71, 2008
|
|
