4UCA
 
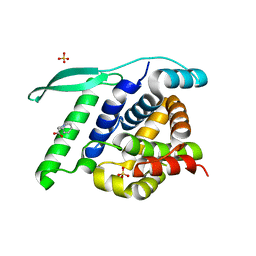 | | N-terminal globular domain of the RSV Nucleoprotein in complex with C- terminal peptide of the Phosphoprotein | | Descriptor: | NUCLEOPROTEIN, PHOSPHOSPROTEIN, SULFATE ION | | Authors: | Ouizougun-Oubari, M, Pereira, N, Tarus, B, Galloux, M, Tortorici, M.-A, Hoos, S, Baron, B, England, P, Bontems, F, Rey, F.A, Eleouet, J.-F, Sizun, C, Slama-Schwok, A, Duquerroy, S. | | Deposit date: | 2014-12-03 | | Release date: | 2015-08-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | A Druggable Pocket at the Nucleocapsid/Phosphoprotein Interaction Site of the Human Respiratory Syncytial Virus.
J.Virol., 89, 2015
|
|
4UC8
 
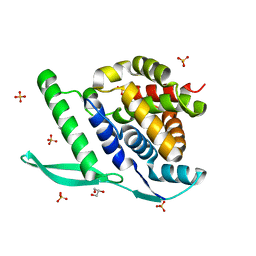 | | N-terminal globular domain of the RSV Nucleoprotein in complex with C- terminal phenylalanine of the Phosphoprotein | | Descriptor: | GLYCEROL, NUCLEOPROTEIN, PHENYLALANINE, ... | | Authors: | Ouizougun-Oubari, M, Pereira, N, Tarus, B, Galloux, M, Tortorici, M.-A, Hoos, S, Baron, B, England, P, Bontems, F, Rey, F.A, Eleouet, J.-F, Sizun, C, Slama-Schwok, A, Duquerroy, S. | | Deposit date: | 2014-12-03 | | Release date: | 2015-08-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Druggable Pocket at the Nucleocapsid/Phosphoprotein Interaction Site of the Human Respiratory Syncytial Virus.
J.Virol., 89, 2015
|
|
9G13
 
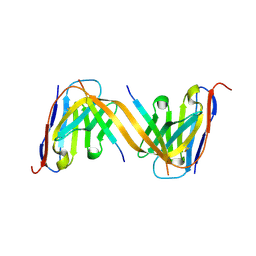 | | VHH H3-2 in complex with Tau C-terminal peptide | | Descriptor: | Isoform Tau-F of Microtubule-associated protein tau, VHH H3-2 | | Authors: | Dupre, E, Landrieu, I, Danis, C, Hanoulle, X, Mortelecque, J. | | Deposit date: | 2024-07-09 | | Release date: | 2024-07-31 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibition of tau neuronal internalization using anti-tau single domain antibodies.
Nat Commun, 16, 2025
|
|
9FQS
 
 | | EGFR Exon20 insertion mutant NPG bound with Compound 39 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3-fluoranyl-2-methoxy-phenyl)amino]-2-[3-[2-[(2R)-1-propanoylpyrrolidin-2-yl]ethynyl]pyridin-4-yl]-1,5,6,7-tetrahydropyrrolo[3,2-c]pyridin-4-one, Epidermal growth factor receptor | | Authors: | Hilbert, B.J, Brooijmans, N, Milgram, B.C, Pagliarini, R.A. | | Deposit date: | 2024-06-17 | | Release date: | 2025-01-29 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | Discovery of STX-721, a Covalent, Potent, and Highly Mutant-Selective EGFR/HER2 Exon20 Insertion Inhibitor for the Treatment of Non-Small Cell Lung Cancer.
J.Med.Chem., 68, 2025
|
|
4V3R
 
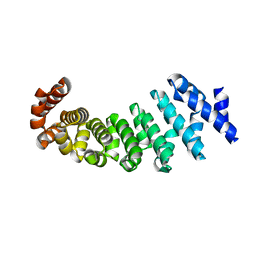 | | Designed armadillo repeat protein with 5 internal repeats, 2nd generation C-cap and 3rd generation N-cap. | | Descriptor: | MAGNESIUM ION, YIII_M5_AII | | Authors: | Reichen, C, Madhurantakam, C, Pluckthun, A, Mittl, P. | | Deposit date: | 2014-10-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of Designed Armadillo-Repeat Proteins Show Propagation of Inter-Repeat Interface Effects
Acta Crystallogr.,Sect.D, 72, 2016
|
|
5KAF
 
 | | RT XFEL structure of Photosystem II in the dark state at 3.0 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Fuller, F, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Waterman, D.G, Evans, G, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.00001 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
4UWN
 
 | | Lysozyme soaked with a ruthenium based CORM with a methione oxide ligand (complex 6b) | | Descriptor: | CARBON MONOXIDE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Santos, M.F.A, Mukhopadhyay, A, Romao, M.J, Romao, C.C, Santos-Silva, T. | | Deposit date: | 2014-08-13 | | Release date: | 2014-12-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | A Contribution to the Rational Design of Ru(Co)3Cl2L Complexes for in Vivo Delivery of Co.
Dalton Trans, 44, 2015
|
|
4UCB
 
 | | N-terminal globular domain of the RSV Nucleoprotein in complex with C- terminal peptide of the Phosphoprotein | | Descriptor: | NUCLEOPROTEIN, PHOSPHOSPROTEIN, SULFATE ION | | Authors: | Ouizougun-Oubari, M, Pereira, N, Tarus, B, Galloux, M, Tortorici, M.-A, Hoos, S, Baron, B, England, P, Bontems, F, Rey, F.A, Eleouet, J.-F, Sizun, C, Slama-Schwok, A, Duquerroy, S. | | Deposit date: | 2014-12-03 | | Release date: | 2015-08-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | A Druggable Pocket at the Nucleocapsid/Phosphoprotein Interaction Site of the Human Respiratory Syncytial Virus.
J.Virol., 89, 2015
|
|
4WF7
 
 | | Crystal structures of trehalose synthase from Deinococcus radiodurans reveal that a closed conformation is involved in the intramolecular isomerization catalysis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wang, Y.L, Chow, S.Y, Lin, Y.T, Hsieh, Y.C, Lee, G.C, Liaw, S.H. | | Deposit date: | 2014-09-13 | | Release date: | 2014-12-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structures of trehalose synthase from Deinococcus radiodurans reveal that a closed conformation is involved in catalysis of the intramolecular isomerization.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
9MX8
 
 | |
4V3O
 
 | | Designed armadillo repeat protein with 5 internal repeats, 2nd generation C-cap and 3rd generation N-cap. | | Descriptor: | ACETATE ION, CALCIUM ION, YIII_M5_AII | | Authors: | Reichen, C, Madhurantakam, C, Pluckthun, A, Mittl, P. | | Deposit date: | 2014-10-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Designed Armadillo-Repeat Proteins Show Propagation of Inter-Repeat Interface Effects
Acta Crystallogr.,Sect.D, 72, 2016
|
|
5XYA
 
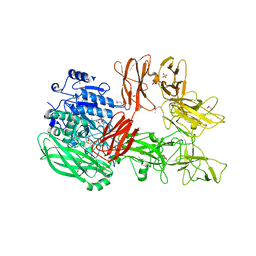 | | Crystal structure of a serine protease from Streptococcus species | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, CALCIUM ION, Chemokine protease C, ... | | Authors: | Jobichen, C, Sivaraman, J. | | Deposit date: | 2017-07-06 | | Release date: | 2018-08-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of ScpC, a virulence protease fromStreptococcus pyogenes, reveals the functional domains and maturation mechanism.
Biochem. J., 475, 2018
|
|
4UJC
 
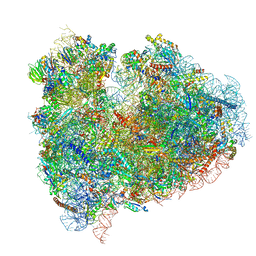 | | mammalian 80S HCV-IRES initiation complex with eIF5B POST-like state | | Descriptor: | 18S RIBOSOMAL RNA, 28S RIBOSOMAL RNA, 40S RIBOSOMAL PROTEIN ES1, ... | | Authors: | Yamamoto, H, Unbehaun, A, Loerke, J, Behrmann, E, Marianne, C, Burger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2014-06-18 | | Release date: | 2014-07-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Structure of the Mammalian 80S Initiation Complex with Initiation Factor 5B on Hcv-Ires RNA.
Nat.Struct.Mol.Biol., 21, 2014
|
|
5XYR
 
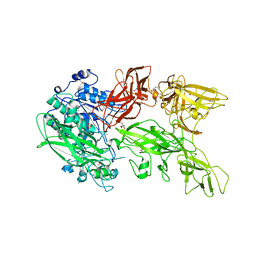 | | Crystal structure of a serine protease from Streptococcus species | | Descriptor: | CALCIUM ION, CHLORIDE ION, Chemokine protease C, ... | | Authors: | Jobichen, C, Sivaraman, J. | | Deposit date: | 2017-07-10 | | Release date: | 2018-08-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of ScpC, a virulence protease fromStreptococcus pyogenes, reveals the functional domains and maturation mechanism.
Biochem. J., 475, 2018
|
|
4UC9
 
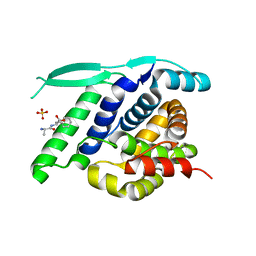 | | N-terminal globular domain of the RSV Nucleoprotein in complex with C- terminal dipeptide of the Phosphoprotein | | Descriptor: | ASPARTIC ACID, NUCLEOPROTEIN, PHENYLALANINE, ... | | Authors: | Ouizougun-Oubari, M, Pereira, N, Tarus, B, Galloux, M, Tortorici, M.-A, Hoos, S, Baron, B, England, P, Bontems, F, Rey, F.A, Eleouet, J.-F, Sizun, C, Slama-Schwok, A, Duquerroy, S. | | Deposit date: | 2014-12-03 | | Release date: | 2015-08-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Druggable Pocket at the Nucleocapsid/Phosphoprotein Interaction Site of the Human Respiratory Syncytial Virus.
J.Virol., 89, 2015
|
|
4UWV
 
 | | Lysozyme soaked with a ruthenium based CORM with a pyridine ligand (complex 8) | | Descriptor: | CARBON MONOXIDE, CHLORIDE ION, LYSOZYME C, ... | | Authors: | Santos, M.F.A, Mukhopadhyay, A, Romao, M.J, Romao, C.C, Santos-Silva, T. | | Deposit date: | 2014-08-14 | | Release date: | 2014-12-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A Contribution to the Rational Design of Ru(Co)3Cl2L Complexes for in Vivo Delivery of Co.
Dalton Trans, 44, 2015
|
|
5YTE
 
 | | Large fragment of DNA Polymerase I from Thermus aquaticus in a closed ternary complex with with natural dT:dATP base pair | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*TP*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | Authors: | Zeng, H, Mondal, M, Song, R.Y, Zhang, J, Xia, B, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2017-11-17 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Unnatural Cytosine Bases Recognized as Thymines by DNA Polymerases by the Formation of the Watson-Crick Geometry.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
4UJD
 
 | | mammalian 80S HCV-IRES initiation complex with eIF5B PRE-like state | | Descriptor: | 18S Ribosomal RNA, 28S Ribosomal RNA, 40S RIBOSOMAL PROTEIN ES1, ... | | Authors: | Yamamoto, H, Unbehaun, A, Loerke, J, Behrmann, E, Marianne, C, Burger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2014-06-18 | | Release date: | 2014-07-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Structure of the Mammalian 80S Initiation Complex with Eif5B on Hcv Ires
Nat.Struct.Mol.Biol., 21, 2014
|
|
5YTD
 
 | | large fragment of DNA Polymerase I from Thermus aquaticus in a closed ternary complex with the natural base pair 5fC:dGTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*(5FC)P*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | Authors: | Zeng, H, Mondal, M, Song, R.Y, Zhang, J, Xia, B, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2017-11-17 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Unnatural Cytosine Bases Recognized as Thymines by DNA Polymerases by the Formation of the Watson-Crick Geometry.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
5YTC
 
 | | Large fragment of DNA Polymerase I from Thermus aquaticus in a closed ternary complex with the unnatural base M-fC pair with dATP in the active site | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*(92F)P*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC)P*(DOC))-3'), ... | | Authors: | Zeng, H, Mondal, M, Song, R.Y, Zhang, J, Xia, B, Yi, C.Q. | | Deposit date: | 2017-11-17 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Unnatural Cytosine Bases Recognized as Thymines by DNA Polymerases by the Formation of the Watson-Crick Geometry.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
5YTF
 
 | | Structure of large fragment of DNA Polymerase I from Thermus aquaticus Host-Guest complex with the unnatural base M-fC pair with dA | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*CP*GP*GP*CP*GP*CP*CP*GP*(92F)P*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | Authors: | Zeng, H, Mondal, M, Song, R.Y, Zhang, J, Xia, B, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2017-11-17 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Unnatural Cytosine Bases Recognized as Thymines by DNA Polymerases by the Formation of the Watson-Crick Geometry.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
5YTG
 
 | | Structure of large fragment of DNA Polymerase I from Thermus aquaticus Host-Guest complex with the unnatural base I-fC pair with dA | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*CP*GP*GP*CP*GP*CP*CP*GP*(94O)P*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | Authors: | Zeng, H, Mondal, M, Song, R.Y, Zhang, J, Xia, B, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2017-11-17 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Unnatural Cytosine Bases Recognized as Thymines by DNA Polymerases by the Formation of the Watson-Crick Geometry.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
5YTH
 
 | | Structure of large fragment of DNA Polymerase I from Thermus aquaticus Host-Guest complex with the unnatural base M-fC pair with dG | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*CP*GP*GP*CP*GP*CP*CP*GP*(92F)P*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*GP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | Authors: | Zeng, H, Mondal, M, Song, R.Y, Zhang, J, Xia, B, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2017-11-17 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Unnatural Cytosine Bases Recognized as Thymines by DNA Polymerases by the Formation of the Watson-Crick Geometry.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
4UYP
 
 | | High resolution structure of the third cohesin ScaC in complex with the ScaB dockerin with a mutation in the N-terminal helix (IN to SI) from Acetivibrio cellulolyticus displaying a type I interaction. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Cameron, K, Alves, V.D, Bule, P, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-09-02 | | Release date: | 2015-04-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Cell-surface Attachment of Bacterial Multienzyme Complexes Involves Highly Dynamic Protein-Protein Anchors.
J. Biol. Chem., 290, 2015
|
|
9MO4
 
 | |
