3CVC
 
 | | Regulation of Protein Function: Crystal Packing Interfaces and Conformational Dimerization | | Descriptor: | COPPER (II) ION, MAGNESIUM ION, Plastocyanin | | Authors: | Crowley, P.B, Matias, P.M, Mi, H, Firbank, S.J, Banfield, M.J, Dennison, C. | | Deposit date: | 2008-04-18 | | Release date: | 2008-07-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Regulation of protein function: crystal packing interfaces and conformational dimerization.
Biochemistry, 47, 2008
|
|
3J2N
 
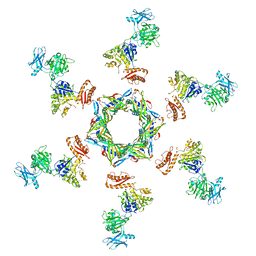 | | The X-ray structure of the gp15 hexamer and the model of the gp18 protein fitted into the cryo-EM reconstruction of the contracted T4 tail | | Descriptor: | Tail connector protein Gp15, Tail sheath protein Gp18 | | Authors: | Fokine, A, Zhang, Z, Kanamaru, S, Bowman, V.D, Aksyuk, A, Arisaka, F, Rao, V.B, Rossmann, M.G. | | Deposit date: | 2012-11-10 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | The molecular architecture of the bacteriophage t4 neck.
J.Mol.Biol., 425, 2013
|
|
3CVJ
 
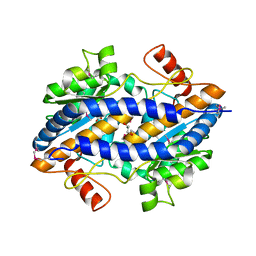 | |
3JWQ
 
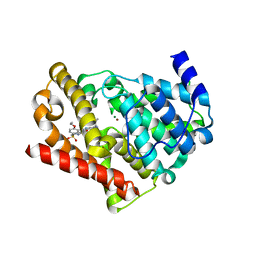 | | Crystal structure of chimeric PDE5/PDE6 catalytic domain complexed with sildenafil | | Descriptor: | 5-{2-ETHOXY-5-[(4-METHYLPIPERAZIN-1-YL)SULFONYL]PHENYL}-1-METHYL-3-PROPYL-1H,6H,7H-PYRAZOLO[4,3-D]PYRIMIDIN-7-ONE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Barren, B, Gakhar, L, Muradov, H, Boyd, K.K, Ramaswamy, S, Artemyev, N.O. | | Deposit date: | 2009-09-18 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structural basis of phosphodiesterase 6 inhibition by the C-terminal region of the gamma-subunit
Embo J., 28, 2009
|
|
3J35
 
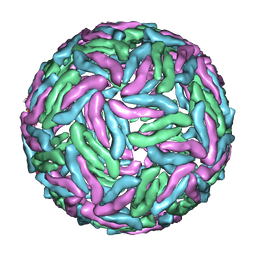 | | Cryo-EM reconstruction of Dengue virus at 37 C | | Descriptor: | envelope protein | | Authors: | Zhang, X.Z, Sheng, J, Plevka, P, Kuhn, R.J, Diamond, M.S, Rossmann, M.G. | | Deposit date: | 2013-02-24 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (35 Å) | | Cite: | Dengue structure differs at the temperatures of its human and mosquito hosts.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3J3S
 
 | | Structural dynamics of the MecA-ClpC complex revealed by cryo-EM | | Descriptor: | Adapter protein MecA 1, Negative regulator of genetic competence ClpC/MecB | | Authors: | Liu, J, Mei, Z, Li, N, Qi, Y, Xu, Y, Shi, Y, Wang, F, Lei, J, Gao, N. | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Structural dynamics of the MecA-ClpC complex: a type II AAA+ protein unfolding machine.
J.Biol.Chem., 288, 2013
|
|
3D6C
 
 | | Crystal structure of Staphylococcal nuclease variant PHS L38E at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Harms, M.J, Schlessman, J.L, Sue, G.R, Garcia-Moreno, E.B. | | Deposit date: | 2008-05-19 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The pK(a) values of acidic and basic residues buried at the same internal location in a protein are governed by different factors.
J.Mol.Biol., 389, 2009
|
|
3J4J
 
 | | Model of full-length T. thermophilus Translation Initiation Factor 2 refined against its cryo-EM density from a 30S Initiation Complex map | | Descriptor: | Translation initiation factor IF-2 | | Authors: | Simonetti, A, Marzi, S, Billas, I.M.L, Tsai, A, Fabbretti, A, Myasnikov, A, Roblin, P, Vaiana, A.C, Hazemann, I, Eiler, D, Steitz, T.A, Puglisi, J.D, Gualerzi, C.O, Klaholz, B.P. | | Deposit date: | 2013-08-26 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Involvement of protein IF2 N domain in ribosomal subunit joining revealed from architecture and function of the full-length initiation factor.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3JX3
 
 | | Structure of rat neuronal nitric oxide synthase D597N/M336V mutant heme domain in complex with N1-{(3'R,4'R)-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-N2-(3'-fluorophenethyl)ethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(3R,4R)-4-[(6-amino-4-methylpyridin-2-yl)methyl]pyrrolidin-3-yl}-N'-[2-(3-fluorophenyl)ethyl]ethane-1,2-diamine, ... | | Authors: | Delker, S.L, Li, H, Poulos, T.L. | | Deposit date: | 2009-09-18 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Unexpected binding modes of nitric oxide synthase inhibitors effective in the prevention of a cerebral palsy phenotype in an animal model.
J.Am.Chem.Soc., 132, 2010
|
|
3JXQ
 
 | | X-Ray structure of r[CGCG(5-fluoro)CG]2 | | Descriptor: | MAGNESIUM ION, r[CGCG(5-fluoro)CG]2 | | Authors: | Adamiak, D.A, Milecki, J, Adamiak, R.W, Rypniewski, W. | | Deposit date: | 2009-09-21 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The hydration and unusual hydrogen bonding in the crystal structure of an RNA duplex containing alternating CG base pairs
New J.Chem., 34, 2010
|
|
3JXV
 
 | |
3J4R
 
 | |
3J63
 
 | | Unified assembly mechanism of ASC-dependent inflammasomes | | Descriptor: | Apoptosis-associated speck-like protein containing a CARD | | Authors: | Lu, A, Magupalli, V.G, Ruan, J, Yin, Q, Atianand, M.K, Vos, M, Schroder, G.F, Fitzgerald, K.A, Wu, H, Egelman, E.H. | | Deposit date: | 2013-12-05 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Unified Polymerization Mechanism for the Assembly of ASC-Dependent Inflammasomes.
Cell(Cambridge,Mass.), 156, 2014
|
|
3D7A
 
 | |
3J6K
 
 | |
3J6L
 
 | | Kinetic and Structural Analysis of Coxsackievirus B3 Receptor Interactions and Formation of the A-particle | | Descriptor: | Coxsackievirus and adenovirus receptor, SULFATE ION | | Authors: | Organtini, L.J, Makhov, A.M, Conway, J.F, Hafenstein, S, Carson, S.D. | | Deposit date: | 2014-03-19 | | Release date: | 2014-04-09 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Kinetic and structural analysis of coxsackievirus b3 receptor interactions and formation of the a-particle.
J.Virol., 88, 2014
|
|
3J6N
 
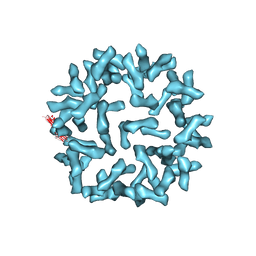 | | Kinetic and Structural Analysis of Coxsackievirus B3 Receptor Interactions and Formation of the A-particle | | Descriptor: | Coxsackie and adenovirus receptor | | Authors: | Organtini, L.J, Makhov, A.M, Conway, J.F, Hafenstein, S, Carson, S.D. | | Deposit date: | 2014-03-19 | | Release date: | 2014-04-09 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Kinetic and structural analysis of coxsackievirus b3 receptor interactions and formation of the a-particle.
J.Virol., 88, 2014
|
|
3J6S
 
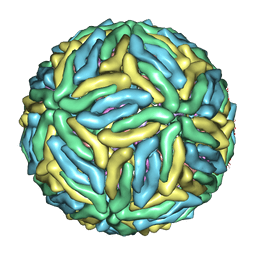 | | Cryo-EM structure of Dengue virus serotype 3 at 28 degrees C | | Descriptor: | envelope protein, membrane protein | | Authors: | Fibriansah, G, Tan, J.L, Smith, S.A, de Alwis, R, Ng, T.-S, Kostyuchenko, V.A, Kukkaro, P, de Silva, A.M, Crowe Jr, J.E, Lok, S.-M. | | Deposit date: | 2014-03-24 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | A highly potent human antibody neutralizes dengue virus serotype 3 by binding across three surface proteins.
Nat Commun, 6, 2015
|
|
3D0R
 
 | | Crystal structure of calG3 from Micromonospora echinospora determined in space group P2(1) | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Protein CalG3 | | Authors: | Bitto, E, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N. | | Deposit date: | 2008-05-02 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and structural insights of the early glycosylation steps in calicheamicin biosynthesis.
Chem.Biol., 15, 2008
|
|
3D1C
 
 | |
3K0N
 
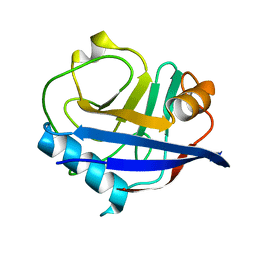 | | Room temperature structure of CypA | | Descriptor: | Cyclophilin A | | Authors: | Fraser, J.S, Alber, T. | | Deposit date: | 2009-09-24 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.391 Å) | | Cite: | Hidden alternative structures of proline isomerase essential for catalysis.
Nature, 462, 2009
|
|
3K0Q
 
 | |
3CI0
 
 | |
3CIM
 
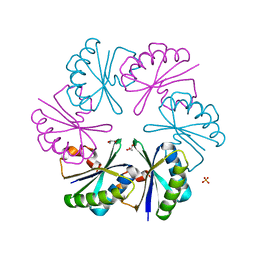 | | Carboxysome shell protein, CcmK2 C-terminal deletion mutant | | Descriptor: | Carbon dioxide-concentrating mechanism protein ccmK homolog 2, GLYCEROL, SULFATE ION | | Authors: | Tanaka, S, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2008-03-11 | | Release date: | 2009-02-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Insights from multiple structures of the shell proteins from the beta-carboxysome.
Protein Sci., 18, 2009
|
|
3K18
 
 | | Tellurium modified DNA-8mer | | Descriptor: | 5'-D(*GP*(UMS)P*GP*(TTI)P*AP*CP*AP*C)-3' | | Authors: | Sheng, J, Hassan, A.E.A, Zhang, W, Huang, Z. | | Deposit date: | 2009-09-25 | | Release date: | 2010-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Synthesis and Structure Study of Oligonucleotides Derivatized with Tellurium Functionality
To be Published
|
|
