5ISH
 
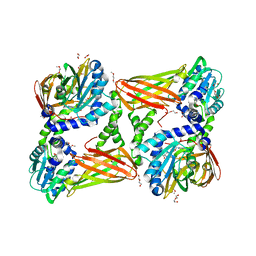 | | Crystal structure of mouse CARM1 in complex with inhibitor SA0765 | | Descriptor: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, 1-METHOXY-2-(2-METHOXYETHOXY)ETHANE, ... | | Authors: | Cura, V, Marechal, N, Mailliot, J, Troffer-Charlier, N, Hassenboehler, P, Wurtz, J.M, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-03-15 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Crystal structure of mouse CARM1 in complex with inhibitor SA0765
To Be Published
|
|
5DNL
 
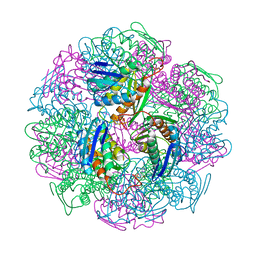 | | Crystal structure of IGPD from Pyrococcus furiosus in complex with (S)-C348 | | Descriptor: | Imidazoleglycerol-phosphate dehydratase, MANGANESE (II) ION, [(2S)-2-hydroxy-3-(1H-1,2,4-triazol-1-yl)propyl]phosphonic acid | | Authors: | Bisson, C, Britton, K.L, Sedelnikova, S.E, Rodgers, H.F, Eadsforth, T.C, Viner, R.C, Hawkes, T.R, Baker, P.J, Rice, D.W. | | Deposit date: | 2015-09-10 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Mirror-Image Packing Provides a Molecular Basis for the Nanomolar Equipotency of Enantiomers of an Experimental Herbicide.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
1WJU
 
 | | Solution structure of N-terminal ubiquitin-like domain of human NEDD8 ultimate buster-1 | | Descriptor: | NEDD8 ultimate buster-1 | | Authors: | Yoneyama, M, Koshiba, S, Tochio, N, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-29 | | Release date: | 2004-11-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of N-terminal ubiquitin-like domain of human NEDD8 ultimate buster-1
To be Published
|
|
5DNX
 
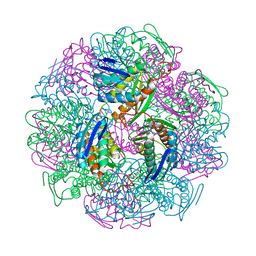 | | Crystal structure of IGPD from Pyrococcus furiosus in complex with (R)-C348 | | Descriptor: | Imidazoleglycerol-phosphate dehydratase, MANGANESE (II) ION, [(2R)-2-hydroxy-3-(1H-1,2,4-triazol-1-yl)propyl]phosphonic acid | | Authors: | Bisson, C, Britton, K.L, Sedelnikova, S.E, Rodgers, H.F, Eadsforth, T.C, Viner, R.C, Hawkes, T.R, Baker, P.J, Rice, D.W. | | Deposit date: | 2015-09-10 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mirror-Image Packing Provides a Molecular Basis for the Nanomolar Equipotency of Enantiomers of an Experimental Herbicide.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
6AQO
 
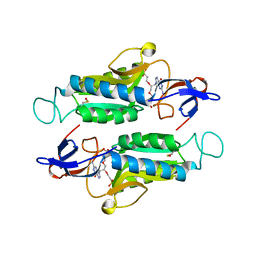 | | Crystal structure of hypoxanthine-guanine-xanthine phosphorybosyltranferase in complex with {[(2S)-3-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)propane-1,2-diyl]bis(oxyethane-2,1-diyl)}bis(phosphonic acid) | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, MAGNESIUM ION, {[(2S)-3-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)propane-1,2-diyl]bis(oxyethane-2,1-diyl)}bis(phosphonic acid) | | Authors: | Teran, D, Gudday, L.W. | | Deposit date: | 2017-08-21 | | Release date: | 2018-03-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Evaluation of the Trypanosoma brucei 6-oxopurine salvage pathway as a potential target for drug discovery.
PLoS Negl Trop Dis, 12, 2018
|
|
4H51
 
 | |
6JB0
 
 | | Crystal structure of ABC transporter alpha-glycoside-binding mutant protein W287A in complex with trehalose | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, ABC transporter, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Gogoi, P. | | Deposit date: | 2019-01-25 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural and thermodynamic correlation illuminates the selective transport mechanism of disaccharide alpha-glycosides through ABC transporter.
Febs J., 287, 2020
|
|
4Q1G
 
 | | Structure and mechanism of a dehydratase/decarboxylase enzyme couple involved in polyketide beta-branching | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Polyketide biosynthesis enoyl-CoA isomerase PksI | | Authors: | Nair, A.V, Race, P.R, Till, M. | | Deposit date: | 2014-04-03 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and mechanism of a dehydratase/decarboxylase enzyme couple involved in polyketide beta-methyl branch incorporation.
Sci Rep, 10, 2020
|
|
4Q1A
 
 | | Human dCK C4S-S74E mutant in complex with UDP and the inhibitor 6 {2-[5-(4-{[(4,6-diaminopyrimidin-2-yl)sulfanyl]methyl}-5-propyl-1,3-thiazol-2-yl)-2-methoxyphenoxy]ethanol} | | Descriptor: | 2-(5-(4-(((4,6-diaminopyrimidin-2-yl)thio)methyl)-5-propylthiazol-2-yl)-2-methoxyphenoxy)ethan-1-ol, Deoxycytidine kinase, URIDINE-5'-DIPHOSPHATE | | Authors: | Nomme, J, Lavie, A. | | Deposit date: | 2014-04-03 | | Release date: | 2015-02-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-guided development of deoxycytidine kinase inhibitors with nanomolar affinity and improved metabolic stability.
J.Med.Chem., 57, 2014
|
|
3KFP
 
 | |
5DSX
 
 | | Crystal structure of Dot1L in complex with inhibitor CPD10 [6'-chloro-1,4-dimethyl-5'-(2-methyl-6-((4-(methylamino)pyrimidin-2-yl)amino)-1H-indol-1-yl)-[3,3'-bipyridin]-2(1H)-one] | | Descriptor: | 6'-chloro-1,4-dimethyl-5'-(2-methyl-6-{[4-(methylamino)pyrimidin-2-yl]amino}-1H-indol-1-yl)-3,3'-bipyridin-2(1H)-one, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Scheufler, C, Gaul, C, Be, C, Moebitz, H. | | Deposit date: | 2015-09-17 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Discovery of Novel Dot1L Inhibitors through a Structure-Based Fragmentation Approach.
Acs Med.Chem.Lett., 7, 2016
|
|
4YAX
 
 | | Crystal structure of TRIM24 PHD-bromodomain complexed with N-[6-(4-methoxyphenoxy)-1,3-dimethyl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl]benzenesulfonamide (5g) | | Descriptor: | GLYCEROL, N-[6-(4-methoxyphenoxy)-1,3-dimethyl-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl]benzenesulfonamide, SULFATE ION, ... | | Authors: | Poncet-Montange, G, Palmer, W, Jones, P. | | Deposit date: | 2015-02-18 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Guided Design of IACS-9571, a Selective High-Affinity Dual TRIM24-BRPF1 Bromodomain Inhibitor.
J.Med.Chem., 59, 2016
|
|
8ZWY
 
 | | Structure-Based Mechanism and Specificity of Human Galactosyltransferase B3GalT5 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-methyl beta-D-galactopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lo, J.M, Ma, C. | | Deposit date: | 2024-06-13 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Mechanism and Specificity of Human Galactosyltransferase beta 3GalT5.
J.Am.Chem.Soc., 147, 2025
|
|
4BJQ
 
 | | Crystal structure of E. coli penicillin binding protein 3, domain V88- S165 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PENICILLIN BINDING PROTEIN TRANSPEPTIDASE DOMAIN PROTEIN, SULFATE ION | | Authors: | Sauvage, E, Joris, M, Herman, R, Kerff, F, Rocaboy, M, Charlier, P. | | Deposit date: | 2013-04-19 | | Release date: | 2014-05-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Penicillin-Binding Protein 3 (Pbp3) from Escherichia Coli.
Plos One, 9, 2014
|
|
8XNF
 
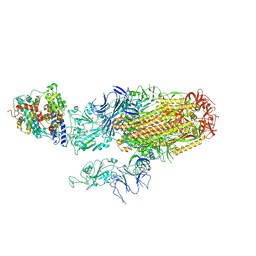 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2.86 spike protein(6P) in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2023-12-29 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
3MNQ
 
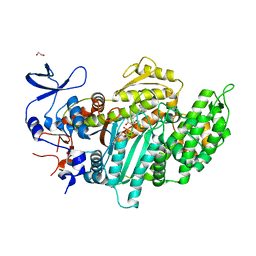 | | Crystal structure of myosin-2 motor domain in complex with ADP-metavanadate and resveratrol | | Descriptor: | 1,2-ETHANEDIOL, ADP METAVANADATE, MAGNESIUM ION, ... | | Authors: | Schneider, J, Taft, M, Backhaus, A, Baruch, P, Fedorov, R, Manstein, D.J. | | Deposit date: | 2010-04-22 | | Release date: | 2011-04-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of resveratrol regulation of myosin activity.
To be Published
|
|
1L5K
 
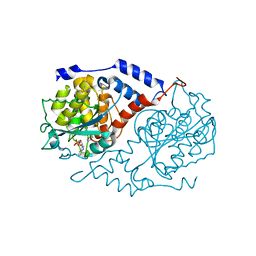 | | Crystal Structure of CobT complexed with N1-(5'-phosphoribosyl)-benzimidazole and nicotinate | | Descriptor: | 1-ALPHA-D-RIBOFURANOSYL-BENZIMIAZOLE-5'-PHOSPHATE, NICOTINIC ACID, Nicotinate-nucleotide--dimethylbenzimidazole phosphoribosyltransferase | | Authors: | Cheong, C.-G, Escalante-Semerena, J, Rayment, I. | | Deposit date: | 2002-03-07 | | Release date: | 2002-09-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies of the L-threonine-O-3-phosphate decarboxylase
(CobD) enzyme from Salmonella enterica: the apo, substrate, and
product-aldimine complexes.
Biochemistry, 41, 2002
|
|
4BGQ
 
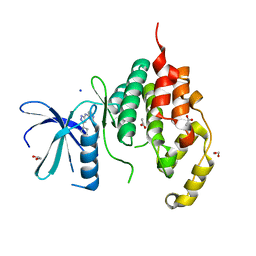 | | Crystal structure of the human CDKL5 kinase domain | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CYCLIN-DEPENDENT KINASE-LIKE 5, ... | | Authors: | Canning, P, Krojer, T, Goubin, S, Mahajan, P, Vollmar, M, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2013-03-28 | | Release date: | 2013-05-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CDKL Family Kinases Have Evolved Distinct Structural Features and Ciliary Function.
Cell Rep, 22, 2018
|
|
2ZB1
 
 | | Crystal structure of P38 in complex with biphenyl amide inhibitor | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase 14, N-(cyclopropylmethyl)-2'-methyl-5'-(5-methyl-1,3,4-oxadiazol-2-yl)biphenyl-4-carboxamide | | Authors: | Somers, D.O. | | Deposit date: | 2007-10-13 | | Release date: | 2008-01-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Biphenyl amide p38 kinase inhibitors 1: Discovery and binding mode
Bioorg.Med.Chem.Lett., 18, 2008
|
|
4QAC
 
 | | X-RAY STRUCTURE OF ACETYLCHOLINE BINDING PROTEIN (ACHBP) IN COMPLEX WITH 4-(4-methylpiperidin-1-yl)-6-(4-(trifluoromethyl)phenyl)pyrimidin-2-amine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(4-methylpiperidin-1-yl)-6-[4-(trifluoromethyl)phenyl]pyrimidin-2-amine, Acetylcholine-binding protein, ... | | Authors: | Kaczanowska, K, Harel, M, Radic, Z, Changeux, J.-P, Finn, M.G, Taylor, P. | | Deposit date: | 2014-05-03 | | Release date: | 2014-07-16 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for cooperative interactions of substituted 2-aminopyrimidines with the acetylcholine binding protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4B5U
 
 | | Crystal structures of divalent metal dependent pyruvate aldolase, HpaI, in complex with pyruvate and succinic semialdehyde | | Descriptor: | 4-HYDROXY-2-OXO-HEPTANE-1,7-DIOATE ALDOLASE, 4-oxobutanoic acid, COBALT (II) ION, ... | | Authors: | Coincon, M, Wang, W, Seah, S.Y.K, Sygusch, J. | | Deposit date: | 2012-08-07 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.913 Å) | | Cite: | Crystal Structure of Reaction Intermediates in Pyruvate Class II Aldolase: Substrate Cleavage, Enolate Stabilization and Substrate Specificity
J.Biol.Chem., 287, 2012
|
|
4B5T
 
 | | Crystal structures of divalent metal dependent pyruvate aldolase, HpaI, in complex with ketobutyrate | | Descriptor: | 2-KETOBUTYRIC ACID, 4-HYDROXY-2-OXO-HEPTANE-1,7-DIOATE ALDOLASE, COBALT (II) ION, ... | | Authors: | Coincon, M, Wang, W, Seah, S.Y.K, Sygusch, J. | | Deposit date: | 2012-08-07 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.923 Å) | | Cite: | Crystal Structure of Reaction Intermediates in Pyruvate Class II Aldolase: Substrate Cleavage, Enolate Stabilization and Substrate Specificity
J.Biol.Chem., 287, 2012
|
|
2W6L
 
 | | The crystal structure at 1.7 A resolution of CobE, a protein from the cobalamin (vitamin B12) biosynthetic pathway | | Descriptor: | COBE, GLYCEROL, SULFATE ION | | Authors: | Vevodova, J, Smith, D, McGoldrick, H, Deery, E, Murzin, A.G, Warren, M.J, Wilson, K.S. | | Deposit date: | 2008-12-18 | | Release date: | 2008-12-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The Crystal Structure at 1.7 A Resolution of Cobe, a Protein from the Cobalamin (Vitamin B12) Biosynthetic Pathway
To be Published
|
|
4B8Y
 
 | | Ferrichrome-bound FhuD2 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FE (III) ION, ... | | Authors: | Malito, E, Bottomley, M.J, Spraggon, G. | | Deposit date: | 2012-08-31 | | Release date: | 2012-11-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Characterization of the Staphylococcus Aureus Virulence Factor and Vaccine Candidate Fhud2.
Biochem.J., 449, 2013
|
|
7FXI
 
 | | Crystal Structure of human FABP4 in complex with rac-(1R,3S)-2,2-dimethyl-3-[(2-phenylphenyl)carbamoyl]cyclopropane-1-carboxylic acid, i.e. SMILES C(=O)(O)[C@@H]1[C@H](C(=O)Nc2ccccc2c2ccccc2)C1(C)C with IC50=0.582 microM | | Descriptor: | (1S,3R)-3-[([1,1'-biphenyl]-2-yl)carbamoyl]-2,2-dimethylcyclopropane-1-carboxylic acid, CHLORIDE ION, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Obst-Sander, U, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
