2H0G
 
 | | Crystal Structure of DsbG T200M mutant | | Descriptor: | SULFATE ION, Thiol:disulfide interchange protein dsbG | | Authors: | Hiniker, A, Heras, B, Martin, J.L, Stuckey, J, Bardwell, J.C.A. | | Deposit date: | 2006-05-15 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Short-circuiting divergent evolution: laboratory evolution of one disulfide isomerase to resemble another
To be Published
|
|
1RL1
 
 | | Solution structure of human Sgt1 CS domain | | Descriptor: | Suppressor of G2 allele of SKP1 homolog | | Authors: | Lee, Y.-T, Jacob, J, Michowski, W, Nowotny, M, Kuznicki, J, Chazin, W.J. | | Deposit date: | 2003-11-24 | | Release date: | 2004-05-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Human Sgt1 Binds HSP90 through the CHORD-Sgt1 Domain and Not the Tetratricopeptide Repeat Domain
J.Biol.Chem., 279, 2004
|
|
2ABH
 
 | | PHOSPHATE-BINDING PROTEIN (RE-REFINED) | | Descriptor: | PHOSPHATE ION, PHOSPHATE-BINDING PROTEIN | | Authors: | Yao, N, Choudhary, A, Ledvina, P.S, Quiocho, F.A. | | Deposit date: | 1995-11-28 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Modulation of a salt link does not affect binding of phosphate to its specific active transport receptor.
Biochemistry, 35, 1996
|
|
1P24
 
 | | Crystal structure of cobalt(II)-d(GGCGCC)2 | | Descriptor: | COBALT (II) ION, DNA (5'-D(*GP*GP*CP*GP*CP*C)-3') | | Authors: | Labiuk, S.L, Delbaere, L.T, Lee, J.S. | | Deposit date: | 2003-04-14 | | Release date: | 2003-12-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Cobalt(II), nickel(II) and zinc(II) do not bind to intra-helical N(7)
guanine positions in the B-form crystal structure of d(GGCGCC)
J.Biol.Inorg.Chem., 8, 2003
|
|
2HPS
 
 | | Crystal structure of coelenterazine-binding protein from Renilla Muelleri | | Descriptor: | C2-HYDROXY-COELENTERAZINE, GLYCEROL, coelenterazine-binding protein with bound coelenterazine | | Authors: | Stepanyuk, G, Liu, Z.J, Vysotski, E.S, Lee, J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-07-17 | | Release date: | 2007-01-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of coelenterazine-binding protein from Renilla muelleri at 1.7 A: why it is not a calcium-regulated photoprotein.
PHOTOCHEM.PHOTOBIOL.SCI., 7, 2008
|
|
1QUJ
 
 | | PHOSPHATE-BINDING PROTEIN MUTANT WITH ASP 137 REPLACED BY GLY COMPLEX WITH CHLORINE AND PHOSPHATE | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, PHOSPHATE-BINDING PROTEIN | | Authors: | Yao, N, Choudhary, A, Ledvina, P.S, Quiocho, F.A. | | Deposit date: | 1995-11-11 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Modulation of a salt link does not affect binding of phosphate to its specific active transport receptor.
Biochemistry, 35, 1996
|
|
1QUL
 
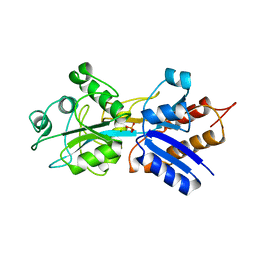 | | PHOSPHATE-BINDING PROTEIN MUTANT WITH ASP 137 REPLACED BY THR COMPLEX WITH CHLORINE AND PHOSPHATE | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, PHOSPHATE-BINDING PROTEIN | | Authors: | Yao, N, Choudhary, A, Ledvina, P.S, Quiocho, F.A. | | Deposit date: | 1995-11-11 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Modulation of a salt link does not affect binding of phosphate to its specific active transport receptor.
Biochemistry, 35, 1996
|
|
1QUK
 
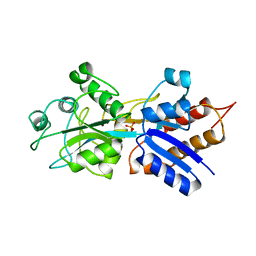 | | PHOSPHATE-BINDING PROTEIN MUTANT WITH ASP 137 REPLACED BY ASN COMPLEX WITH PHOSPHATE | | Descriptor: | PHOSPHATE ION, PHOSPHATE-BINDING PROTEIN | | Authors: | Yao, N, Choudhary, A, Ledvina, P.S, Quiocho, F.A. | | Deposit date: | 1995-11-11 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Modulation of a salt link does not affect binding of phosphate to its specific active transport receptor.
Biochemistry, 35, 1996
|
|
1PZZ
 
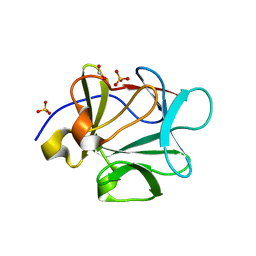 | | Crystal structure of FGF-1, V51N mutant | | Descriptor: | FORMIC ACID, Heparin-binding growth factor 1, SULFATE ION | | Authors: | Kim, J, Blaber, M. | | Deposit date: | 2003-07-14 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sequence swapping does not result in conformation swapping for the beta4/beta5 and beta8/beta9 beta-hairpin turns in human acidic fibroblast growth factor
Protein Sci., 14, 2005
|
|
2R0W
 
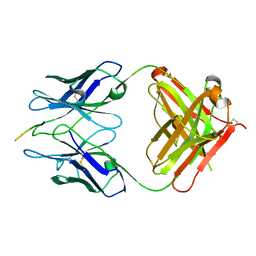 | | PFA2 FAB complexed with Abeta1-8 | | Descriptor: | Amyloid beta peptide fragment, IgG2a Fab fragment heavy chain, Fd portion, ... | | Authors: | Gardberg, A.S, Dealwis, C. | | Deposit date: | 2007-08-21 | | Release date: | 2007-10-16 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Molecular basis for passive immunotherapy of Alzheimer's disease
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2KSM
 
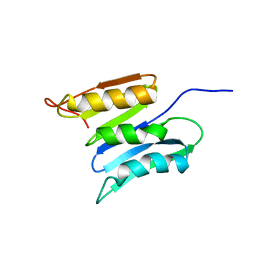 | | Central B domain of Rv0899 from Mycobacterium tuberculosis | | Descriptor: | MYCOBACTERIUM TUBERCULOSIS RV0899/MT0922/OmpATb | | Authors: | Teriete, P, Yao, Y, Kolodzik, A, Yu, J, Song, H, Niederweis, M, Marassi, F.M. | | Deposit date: | 2010-01-07 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Mycobacterium tuberculosis Rv0899 adopts a mixed alpha/beta-structure and does not form a transmembrane beta-barrel.
Biochemistry, 49, 2010
|
|
2MGX
 
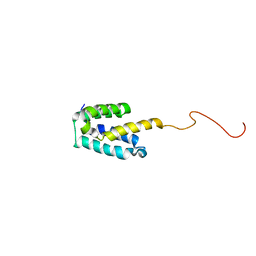 | | NMR structure of SRA1p C-terminal domain | | Descriptor: | Steroid receptor RNA activator 1 | | Authors: | Bilinovich, S.M, Davis, C.M, Morris, D.L, Ray, L.A, Prokop, J.W, Buchan, G.J, Leeper, T.C. | | Deposit date: | 2013-11-10 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The C-Terminal Domain of SRA1p Has a Fold More Similar to PRP18 than to an RRM and Does Not Directly Bind to the SRA1 RNA STR7 Region.
J.Mol.Biol., 426, 2014
|
|
1MQH
 
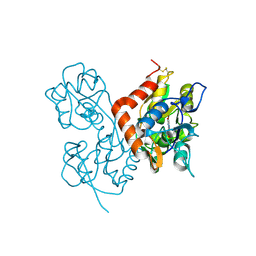 | | Crystal Structure of the GluR2 Ligand Binding Core (S1S2J) in Complex with Bromo-Willardiine at 1.8 Angstroms Resolution | | Descriptor: | 2-AMINO-3-(5-BROMO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, glutamate receptor 2 | | Authors: | Jin, R, Banke, T.G, Mayer, M.L, Traynelis, S.F, Gouaux, E. | | Deposit date: | 2002-09-16 | | Release date: | 2003-08-05 | | Last modified: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for partial agonist action at ionotropic glutamate receptors
Nat.Neurosci., 6, 2003
|
|
1MQI
 
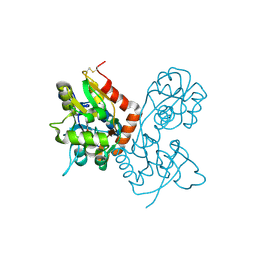 | | Crystal Structure of the GluR2 Ligand Binding Core (S1S2J) in Complex with Fluoro-Willardiine at 1.35 Angstroms Resolution | | Descriptor: | 2-AMINO-3-(5-FLUORO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, glutamate receptor 2 | | Authors: | Jin, R, Banke, T.G, Mayer, M.L, Traynelis, S.F, Gouaux, E. | | Deposit date: | 2002-09-16 | | Release date: | 2003-08-05 | | Last modified: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural basis for partial agonist action at ionotropic glutamate receptors
Nat.Neurosci., 6, 2003
|
|
1MQJ
 
 | | Crystal structure of the GluR2 ligand binding core (S1S2J) in complex with willardiine at 1.65 angstroms resolution | | Descriptor: | 2-AMINO-3-(2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, ZINC ION, glutamate receptor 2 | | Authors: | Jin, R, Banke, T.G, Mayer, M.L, Traynelis, S.F, Gouaux, E. | | Deposit date: | 2002-09-16 | | Release date: | 2003-08-05 | | Last modified: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for partial agonist action at ionotropic glutamate receptors
Nat.Neurosci., 6, 2003
|
|
1MQG
 
 | | Crystal Structure of the GluR2 Ligand Binding Core (S1S2J) in Complex with Iodo-Willardiine at 2.15 Angstroms Resolution | | Descriptor: | 2-AMINO-3-(5-IODO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Jin, R, Banke, T.G, Mayer, M.L, Traynelis, S.F, Gouaux, E. | | Deposit date: | 2002-09-16 | | Release date: | 2003-08-05 | | Last modified: | 2017-05-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for partial agonist action at ionotropic glutamate receptors
Nat.Neurosci., 6, 2003
|
|
3E6F
 
 | | MHC CLASS I H-2Dd Heavy chain complexed with Beta-2 Microglobulin and a variant peptide, PA9, from the Human immunodeficiency virus (BaL) envelope glycoprotein 120 | | Descriptor: | BETA-2 MICROGLOBULIN, Envelope glycoprotein 9-residue peptide, H-2 class I histocompatibility antigen, ... | | Authors: | Wang, R, Natarajan, K, Robinson, H, Margulies, D.H. | | Deposit date: | 2008-08-15 | | Release date: | 2009-08-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Different vaccine vectors delivering the same antigen elicit CD8+ T cell responses with distinct clonotype and epitope specificity
J.Immunol., 183, 2009
|
|
3E6H
 
 | | MHC CLASS I H-2Dd heavy chain complexed with Beta-2 Microglobulin and a variant peptide, PI10, from the human immunodeficiency virus (BaL) envelope glycoprotein 120 | | Descriptor: | Envelope glycoprotein 10-residue peptide, H-2 class I histocompatibility antigen, D-D alpha chain, ... | | Authors: | Wang, R, Natarajan, K, Robinson, H, Margulies, D.H. | | Deposit date: | 2008-08-15 | | Release date: | 2009-08-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Different vaccine vectors delivering the same antigen elicit CD8+ T cell responses with distinct clonotype and epitope specificity
J.Immunol., 183, 2009
|
|
4WHX
 
 | | X-ray Crystal Structure of an Amino Acid Aminotransferase from Burkholderia pseudomallei Bound to the Co-factor Pyridoxal Phosphate | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, Branched-chain-amino-acid transaminase | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID), Fairman, J.W, Dranow, D.M, Taylor, B.M, Lorimer, D, Edwards, T.E. | | Deposit date: | 2014-09-23 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray Crystal Structure of an Amino Acid Aminotransferase from Burkholderia pseudomallei Bound to the Co-factor Pyridoxal Phosphate
to be published
|
|
2H0H
 
 | | Crystal Structure of DsbG K113E mutant | | Descriptor: | SULFATE ION, Thiol:disulfide interchange protein dsbG | | Authors: | Hiniker, A, Heras, B, Martin, J.L, Stuckey, J, Bardwell, J.C.A. | | Deposit date: | 2006-05-15 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Short-circuiting divergent evolution: laboratory evolution of one disulfide isomerase to resemble another
To be Published
|
|
4OVR
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM XANTHOBACTER AUTOTROPHICUS PY2, TARGET EFI-510329, WITH BOUND BETA-D-GALACTURONATE | | Descriptor: | CHLORIDE ION, TRAP dicarboxylate transporter, DctP subunit, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-12-12 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4P1L
 
 | | Crystal structure of a trap periplasmic solute binding protein from chromohalobacter salexigens dsm 3043 (csal_2479), target EFI-510085, with bound d-glucuronate, spg i213 | | Descriptor: | GLYCEROL, SULFATE ION, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-02-26 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4P3L
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM CHROMOHALOBACTER SALEXIGENS DSM 3043 (Csal_2479), TARGET EFI-510085, WITH BOUND GLUCURONATE, SPG P6122 | | Descriptor: | CHLORIDE ION, TRAP dicarboxylate transporter, DctP subunit, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-03-09 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4P1E
 
 | | Crystal structure of a trap periplasmic solute binding protein from escherichia fergusonii (efer_1530), target EFI-510119, apo open structure, phased with iodide | | Descriptor: | IODIDE ION, TRAP dicarboxylate transporter, DctP subunit | | Authors: | Vetting, M.W, Al Obaidi, N.F, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-02-26 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4P47
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM OCHROBACTRUM ANTHROPI (Oant_4429), TARGET EFI-510151, C-TERMIUS BOUND IN LIGAND BINDING POCKET | | Descriptor: | TRAP dicarboxylate transporter, DctP subunit | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-03-12 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
