4MQ5
 
 | |
4LCW
 
 | | The structure of human MAIT TCR in complex with MR1-K43A-RL-6-Me-7OH | | Descriptor: | 1-deoxy-1-(7-hydroxy-6-methyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Patel, O, Rossjohn, J. | | Deposit date: | 2013-06-24 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Antigen-loaded MR1 tetramers define T cell receptor heterogeneity in mucosal-associated invariant T cells.
J.Exp.Med., 210, 2013
|
|
6MCX
 
 | | INFLUENZA VIRUS NEURAMINIDASE SUBTYPE N9 (TERN) RECOMBINANT HEAD DOMAIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Streltsov, V.A, Schmidt, P, McKimm-Breschkin, J. | | Deposit date: | 2018-09-02 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of an Influenza A virus N9 neuraminidase with a tetrabrachion-domain stalk.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
8USW
 
 | | CNQX-bound GluN1a-3A NMDA receptor | | Descriptor: | 7-nitro-2,3-dioxo-1,2,3,4-tetrahydroquinoxaline-6-carbonitrile, Glutamate receptor ionotropic, NMDA 1, ... | | Authors: | Michalski, K, Furukawa, H. | | Deposit date: | 2023-10-30 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | Structure and function of GluN1-3A NMDA receptor excitatory glycine receptor channel.
Sci Adv, 10, 2024
|
|
6ABI
 
 | | The apo-structure of D-lactate dehydrogenase from Fusobacterium nucleatum | | Descriptor: | D-lactate dehydrogenase, GLYCEROL, SULFATE ION | | Authors: | Furukawa, N, Miyanaga, A, Nakajima, M, Taguchi, H. | | Deposit date: | 2018-07-21 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Sequential Allosteric Transitions in Tetrameric d-Lactate Dehydrogenases from Three Gram-Negative Bacteria
Biochemistry, 57, 2018
|
|
6ABJ
 
 | | The apo-structure of D-lactate dehydrogenase from Pseudomonas aeruginosa | | Descriptor: | ACETATE ION, D-lactate dehydrogenase (Fermentative) | | Authors: | Furukawa, N, Miyanaga, A, Nakajima, M, Taguchi, H. | | Deposit date: | 2018-07-21 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural Basis of Sequential Allosteric Transitions in Tetrameric d-Lactate Dehydrogenases from Three Gram-Negative Bacteria
Biochemistry, 57, 2018
|
|
4HQ9
 
 | |
6P6U
 
 | |
6FL9
 
 | |
4MPU
 
 | | Human beta-tryptase co-crystal structure with (6S,8R)-N,N'-bis[3-({4-[3-(aminomethyl)phenyl]piperidin-1-yl}carbonyl)phenyl]-8-hydroxy-6-(1-hydroxycyclobutyl)-5,7-dioxaspiro[3.4]octane-6,8-dicarboxamide | | Descriptor: | (6S,8R)-N,N'-bis[3-({4-[3-(aminomethyl)phenyl]piperidin-1-yl}carbonyl)phenyl]-8-hydroxy-6-(1-hydroxycyclobutyl)-5,7-dioxaspiro[3.4]octane-6,8-dicarboxamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, ... | | Authors: | White, A, Stein, A.J, Suto, R.K. | | Deposit date: | 2013-09-13 | | Release date: | 2015-03-18 | | Last modified: | 2019-09-25 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Target-Directed Self-Assembly of Homodimeric Drugs Against beta-Tryptase.
Acs Med.Chem.Lett., 9, 2018
|
|
4OO2
 
 | | Streptomyces globisporus C-1027 FAD dependent (S)-3-chloro-β-tyrosine-S-SgcC2 C-5 hydroxylase SgcC apo form | | Descriptor: | CALCIUM ION, Chlorophenol-4-monooxygenase, GLYCEROL | | Authors: | Cao, H, Xu, W, Bingman, C.A, Lohman, J.R, Yennamalli, R, Shen, B, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-01-29 | | Release date: | 2014-02-12 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Crystal Structures of SgcE6 and SgcC, the Two-Component Monooxygenase That Catalyzes Hydroxylation of a Carrier Protein-Tethered Substrate during the Biosynthesis of the Enediyne Antitumor Antibiotic C-1027 in Streptomyces globisporus.
Biochemistry, 55, 2016
|
|
4X8V
 
 | | FACTOR VIIA IN COMPLEX WITH THE INHIBITOR (methyl {3-[(2R)-1-{(2R)-2-(3,4-dimethoxyphenyl)-2-[(1-oxo-1,2,3,4-tetrahydroisoquinolin-7-yl)amino]acetyl}pyrrolidin-2-yl]-4-(propan-2-ylsulfonyl)phenyl}carbamate) | | Descriptor: | CALCIUM ION, Factor VIIa (Heavy Chain), Factor VIIa (Light Chain), ... | | Authors: | Wei, A. | | Deposit date: | 2014-12-10 | | Release date: | 2015-03-25 | | Last modified: | 2015-04-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Novel P1 Groups for Coagulation Factor VIIa Inhibition Using Fragment-Based Screening.
J.Med.Chem., 58, 2015
|
|
8FO1
 
 | |
4YVQ
 
 | |
5DED
 
 | | Crystal structure of the small alarmone synthethase 1 from Bacillus subtilis bound to its product pppGpp | | Descriptor: | GTP pyrophosphokinase YjbM, MAGNESIUM ION, guanosine 5'-(tetrahydrogen triphosphate) 3'-(trihydrogen diphosphate) | | Authors: | Steinchen, W, Schuhmacher, J.S, Altegoer, F, Bange, G. | | Deposit date: | 2015-08-25 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.942 Å) | | Cite: | Catalytic mechanism and allosteric regulation of an oligomeric (p)ppGpp synthetase by an alarmone.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4AAY
 
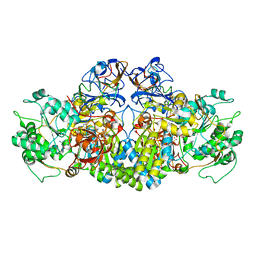 | | Crystal Structure of the arsenite oxidase protein complex from Rhizobium species strain NT-26 | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, AROA, AROB, ... | | Authors: | Oke, M, Santini, J.M, Naismith, J.H. | | Deposit date: | 2011-12-05 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Respiratory Arsenite Oxidase: Structure and the Role of Residues Surrounding the Rieske Cluster.
Plos One, 8, 2013
|
|
4H2Y
 
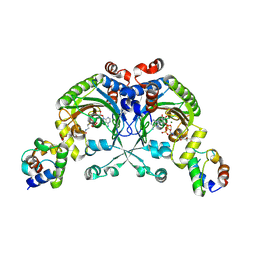 | | Crystal structure of engineered Bradyrhizobium japonicum glycine:[carrier protein] ligase complexed with carrier protein from Agrobacterium tumefaciens and ATP | | Descriptor: | 4'-PHOSPHOPANTETHEINE, ADENOSINE-5'-TRIPHOSPHATE, Amino acid--[acyl-carrier-protein] ligase 1, ... | | Authors: | Luic, M, Weygand-Durasevic, I, Ivic, N, Mocibob, M. | | Deposit date: | 2012-09-13 | | Release date: | 2013-03-06 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Adaptation of aminoacyl-tRNA synthetase catalytic core to carrier protein aminoacylation.
Structure, 21, 2013
|
|
4H2S
 
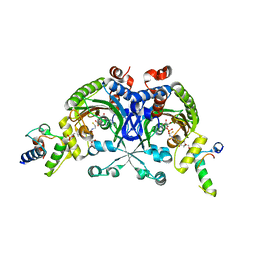 | | Crystal structure of Bradyrhizobium japonicum glycine:[carrier protein] ligase complexed with cognate carrier protein and AMP | | Descriptor: | 4'-PHOSPHOPANTETHEINE, ACETATE ION, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Luic, M, Weygand-Durasevic, I, Ivic, N, Mocibob, M. | | Deposit date: | 2012-09-13 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Adaptation of aminoacyl-tRNA synthetase catalytic core to carrier protein aminoacylation.
Structure, 21, 2013
|
|
8IE2
 
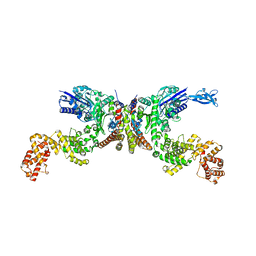 | | Crystal structure of Lactiplantibacillus plantarum GlyRS | | Descriptor: | Glycine--tRNA ligase alpha subunit, Glycine--tRNA ligase beta subunit | | Authors: | Yamashita, S, Tomita, K. | | Deposit date: | 2023-02-15 | | Release date: | 2023-06-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Mechanism of tRNA recognition by heterotetrameric glycyl-tRNA synthetase from lactic acid bacteria.
J.Biochem., 174, 2023
|
|
4H2X
 
 | | Crystal structure of engineered Bradyrhizobium japonicum glycine:[carrier protein] ligase complexed with carrier protein from Agrobacterium tumefaciens and an analogue of glycyl adenylate | | Descriptor: | 4'-PHOSPHOPANTETHEINE, 5'-O-(glycylsulfamoyl)adenosine, Amino acid--[acyl-carrier-protein] ligase 1, ... | | Authors: | Luic, M, Weygand-Durasevic, I, Ivic, N, Mocibob, M. | | Deposit date: | 2012-09-13 | | Release date: | 2013-03-06 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Adaptation of aminoacyl-tRNA synthetase catalytic core to carrier protein aminoacylation.
Structure, 21, 2013
|
|
4H2U
 
 | | Crystal structure of Bradyrhizobium japonicum glycine:[carrier protein] ligase complexed with cognate carrier protein and ATP | | Descriptor: | 4'-PHOSPHOPANTETHEINE, ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Luic, M, Weygand-Durasevic, I, Ivic, N, Mocibob, M. | | Deposit date: | 2012-09-13 | | Release date: | 2013-03-06 | | Last modified: | 2013-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Adaptation of aminoacyl-tRNA synthetase catalytic core to carrier protein aminoacylation.
Structure, 21, 2013
|
|
4GO0
 
 | |
4GNZ
 
 | |
4GO2
 
 | |
4PXI
 
 | | Elucidation of the Structural and Functional Mechanism of Action of the TetR Family Protein, CprB from S. coelicolor A3(2) | | Descriptor: | CprB, DNA (5'-D(*AP*CP*AP*TP*AP*CP*GP*GP*GP*AP*CP*GP*CP*CP*CP*CP*GP*TP*TP*TP*AP*T)-3'), DNA (5'-D(*AP*TP*AP*AP*AP*CP*GP*GP*GP*GP*CP*GP*TP*CP*CP*CP*GP*TP*AP*TP*GP*T)-3') | | Authors: | Hussain, B, Ruchika, B, Aruna, B, Ruchi, A. | | Deposit date: | 2014-03-24 | | Release date: | 2014-07-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and functional basis of transcriptional regulation by TetR family protein CprB from S. coelicolor A3(2)
Nucleic Acids Res., 42, 2014
|
|
