7QTU
 
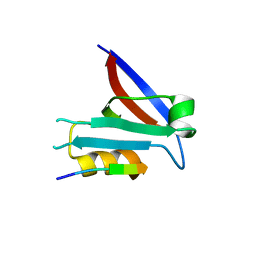 | |
2BIE
 
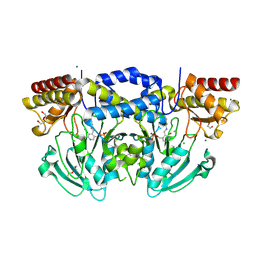 | | Radiation damage of the Schiff base in phosphoserine aminotransferase (structure H) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Dubnovitsky, A.P, Ravelli, R.B.G, Popov, A.N, Papageorgiou, A.C. | | Deposit date: | 2005-01-21 | | Release date: | 2005-05-19 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Strain Relief at the Active Site of Phosphoserine Aminotransferase Induced by Radiation Damage.
Protein Sci., 14, 2005
|
|
1X8K
 
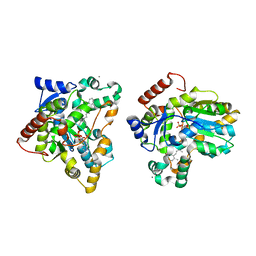 | | Crystal structure of retinol dehydratase in complex with anhydroretinol and inactive cofactor PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, ANHYDRORETINOL, CALCIUM ION, ... | | Authors: | Pakhomova, S, Buck, J, Newcomer, M.E. | | Deposit date: | 2004-08-18 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The structures of the unique sulfotransferase retinol dehydratase with product and inhibitors provide insight into enzyme mechanism and inhibition.
Protein Sci., 14, 2005
|
|
1XH0
 
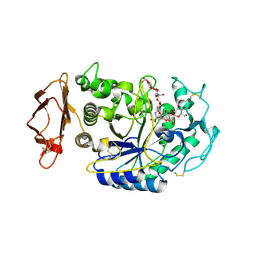 | | Structure of the N298S variant of human pancreatic alpha-amylase complexed with acarbose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACARBOSE DERIVED HEXASACCHARIDE, Alpha-amylase, ... | | Authors: | Maurus, R, Begum, A, Kuo, H.H, Racaza, A, Numao, S, Overall, C.M, Withers, S.G, Brayer, G.D. | | Deposit date: | 2004-09-17 | | Release date: | 2005-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and mechanistic studies of chloride induced activation of human pancreatic alpha-amylase
PROTEIN SCI., 14, 2005
|
|
1XGZ
 
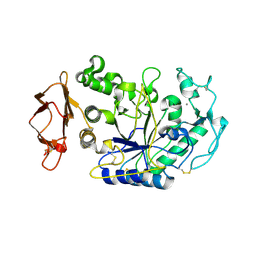 | | Structure of the N298S variant of human pancreatic alpha-amylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase, pancreatic, ... | | Authors: | Maurus, R, Begum, A, Kuo, H.H, Racaza, A, Numao, S, Overall, C.M, Withers, S.G, Brayer, G.D. | | Deposit date: | 2004-09-17 | | Release date: | 2005-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and mechanistic studies of chloride induced activation of human pancreatic alpha-amylase
PROTEIN SCI., 14, 2005
|
|
4JHP
 
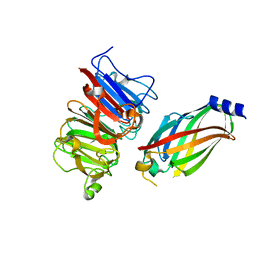 | | The crystal structure of the RPGR RCC1-like domain in complex with PDE6D | | Descriptor: | Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta, X-linked retinitis pigmentosa GTPase regulator | | Authors: | Waetzlich, D, Vetter, I, Wittinghofer, A, Ismail, S. | | Deposit date: | 2013-03-05 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The interplay between RPGR, PDE-delta and Arl2/3 regulate the ciliary targeting of farnesylated cargo.
Embo Rep., 14, 2013
|
|
7QFC
 
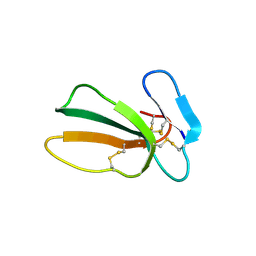 | | Crystal structure of cytotoxin 13 from Naja naja, orthorhombic form | | Descriptor: | Cytotoxin 13 | | Authors: | Samygina, V.R, Dubova, K.M, Bourenkov, G, Utkin, Y.N, Dubovskii, P.V. | | Deposit date: | 2021-12-05 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Variability in the Spatial Structure of the Central Loop in Cobra Cytotoxins Revealed by X-ray Analysis and Molecular Modeling.
Toxins, 14, 2022
|
|
7QHI
 
 | | Crystal structure of cytotoxin 13 from Naja naja, hexagonal form | | Descriptor: | Cytotoxin 13 | | Authors: | Samygina, V.R, Dubova, K.M, Bourenkov, G, Utkin, Y.N, Dubovskii, P.V. | | Deposit date: | 2021-12-12 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Variability in the Spatial Structure of the Central Loop in Cobra Cytotoxins Revealed by X-ray Analysis and Molecular Modeling.
Toxins, 14, 2022
|
|
4JKW
 
 | |
1XFL
 
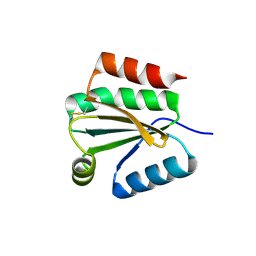 | | Solution Structure of Thioredoxin h1 from Arabidopsis Thaliana | | Descriptor: | Thioredoxin h1 | | Authors: | Peterson, F.C, Lytle, B.L, Sampath, S, Vinarov, D, Tyler, E, Shahan, M, Markley, J.L, Volkman, B.F, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-09-15 | | Release date: | 2004-09-28 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of thioredoxin h1 from Arabidopsis thaliana.
Protein Sci., 14, 2005
|
|
4JLQ
 
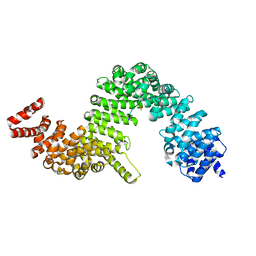 | | Crystal structure of human Karyopherin-beta2 bound to the PY-NLS of Saccharomyces cerevisiae NAB2 | | Descriptor: | Nuclear polyadenylated RNA-binding protein NAB2, Transportin-1 | | Authors: | Sampathkumar, P, Gizzi, A, Rout, M.P, Chook, Y.M, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Nucleocytoplasmic Transport: a Target for Cellular Control (NPCXstals) | | Deposit date: | 2013-03-12 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of human Karyopherin beta 2 bound to the PY-NLS of Saccharomyces cerevisiae Nab2.
J.Struct.Funct.Genom., 14, 2013
|
|
1XJI
 
 | | Bacteriorhodopsin crystallized in bicelles at room temperature | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Bacteriorhodopsin, DECANE, ... | | Authors: | Faham, S, Boulting, G.L, Massey, E.A, Yohannan, S, Yang, D, Bowie, J.U. | | Deposit date: | 2004-09-23 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallization of bacteriorhodopsin from bicelle formulations at room temperature
Protein Sci., 14, 2005
|
|
2BZT
 
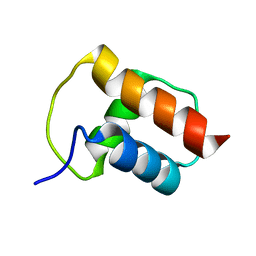 | | NMR structure of the bacterial protein YFHJ from E. coli | | Descriptor: | PROTEIN ISCX | | Authors: | Pastore, C, Kelly, G, Adinolfi, S, Mc Cormick, J.E, Pastore, A. | | Deposit date: | 2005-08-22 | | Release date: | 2006-12-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | YfhJ, a molecular adaptor in iron-sulfur cluster formation or a frataxin-like protein?
Structure, 14, 2006
|
|
2JT0
 
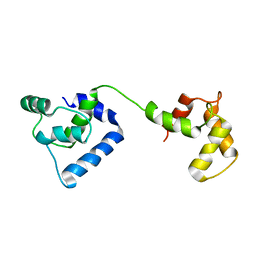 | | Solution structure of F104W cardiac troponin C | | Descriptor: | Troponin C, slow skeletal and cardiac muscles | | Authors: | Wang, X, Mercier, P, Letourneau, P.-J, Sykes, B.D. | | Deposit date: | 2007-07-17 | | Release date: | 2008-05-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Effects of Phe-to-Trp mutation and fluorotryptophan incorporation on the solution structure of cardiac troponin C, and analysis of its suitability as a potential probe for in situ NMR studies.
Protein Sci., 14, 2005
|
|
4JC1
 
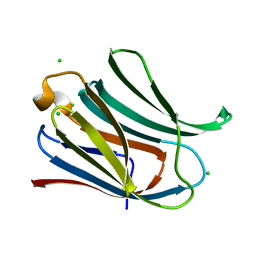 | |
7Q64
 
 | | Cryo-em structure of the Nup98 fibril polymorph 1 | | Descriptor: | Nuclear pore complex protein Nup98 | | Authors: | Ibanez de Opakua, A, Geraets, J.A, Frieg, B, Dienemann, C, Savastano, A, Rankovic, M, Cima-Omori, M.-S, Schroeder, G.F, Zweckstetter, M. | | Deposit date: | 2021-11-05 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Molecular interactions of FG nucleoporin repeats at high resolution.
Nat.Chem., 14, 2022
|
|
1ZL3
 
 | | Coupling of active site motions and RNA binding | | Descriptor: | 5'-R(*GP*GP*CP*AP*AP*CP*GP*GP*UP*(FLO) UP*CP*GP*AP*UP*CP*CP*CP*GP*UP*UP*GP*C)-3', SULFATE ION, tRNA pseudouridine synthase B | | Authors: | Hoang, C, Hamilton, C.S, Mueller, E.G, Ferre-D'Amare, A.R. | | Deposit date: | 2005-05-05 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Precursor complex structure of pseudouridine synthase TruB suggests coupling of active site perturbations to an RNA-sequestering peripheral protein domain
Protein Sci., 14, 2005
|
|
7Q65
 
 | | Cryo-em structure of the Nup98 fibril polymorph 2 | | Descriptor: | Nuclear pore complex protein Nup98 | | Authors: | Ibanez de Opakua, A, Geraets, J.A, Frieg, B, Dienemann, C, Savastano, A, Rankovic, M, Cima-Omori, M.-S, Schroeder, G.F, Zweckstetter, M. | | Deposit date: | 2021-11-05 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Molecular interactions of FG nucleoporin repeats at high resolution.
Nat.Chem., 14, 2022
|
|
1ZA4
 
 | |
4JIP
 
 | |
7Q67
 
 | | Cryo-em structure of the Nup98 fibril polymorph 4 | | Descriptor: | Nuclear pore complex protein Nup98 | | Authors: | Ibanez de Opakua, A, Geraets, J.A, Frieg, B, Dienemann, C, Savastano, A, Rankovic, M, Cima-Omori, M.-S, Schroeder, G.F, Zweckstetter, M. | | Deposit date: | 2021-11-05 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Molecular interactions of FG nucleoporin repeats at high resolution.
Nat.Chem., 14, 2022
|
|
2AU1
 
 | | Crystal Structure of group A Streptococcus MAC-1 orthorhombic form | | Descriptor: | BETA-MERCAPTOETHANOL, IgG-degrading protease | | Authors: | Agniswamy, J, Nagiec, M.J, Liu, M, Schuck, P, Musser, J.M, Sun, P.D. | | Deposit date: | 2005-08-26 | | Release date: | 2006-02-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of group a streptococcus mac-1: insight into dimer-mediated specificity for recognition of human IgG.
Structure, 14, 2006
|
|
2CGX
 
 | | Identification of chemically diverse Chk1 inhibitors by receptor- based virtual screening | | Descriptor: | 2-[(6-AMINO-7H-PURIN-8-YL)THIO]ACETAMIDE, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Foloppe, N, Fisher, L.M, Howes, R, Potter, A, Robertson, A.G.S, Surgenor, A.E. | | Deposit date: | 2006-03-09 | | Release date: | 2006-04-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of Chemically Diverse Chk1 Inhibitors by Receptor-Based Virtual Screening.
Bioorg.Med.Chem., 14, 2006
|
|
2CI4
 
 | | Crystal Structure of Dimethylarginine dimethylaminohydrolase I crystal form II | | Descriptor: | NG, NG-DIMETHYLARGININE DIMETHYLAMINOHYDROLASE 1 | | Authors: | Frey, D, Braun, O, Briand, C, Vasak, M, Grutter, M.G. | | Deposit date: | 2006-03-17 | | Release date: | 2006-05-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the Mammalian Nos Regulator Dimethylarginine Dimethylaminohydrolase: A Basis for the Design of Specific Inhibitors.
Structure, 14, 2006
|
|
7QDD
 
 | |
