5TNB
 
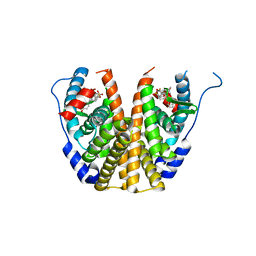 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S,L536S) in Complex with the OBHS-BSC, 4-bromophenyl (1R,2R,4S)-6-(4-(2-(dimethylamino)ethoxy)phenyl)-5-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate | | Descriptor: | 4-bromophenyl (1S,2R,4S)-6-{4-[2-(dimethylamino)ethoxy]phenyl}-5-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate, Estrogen receptor | | Authors: | Nwachukwu, J.C, Sharma, N, Carlson, K.E, Srinivasan, S, Sharma, A, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-02-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Exploring the Structural Compliancy versus Specificity of the Estrogen Receptor Using Isomeric Three-Dimensional Ligands.
ACS Chem. Biol., 12, 2017
|
|
5ESS
 
 | | Crystal Structure of M. tuberculosis MenD bound to Mg2+ and covalent intermediate I (a ThDP and decarboxylated 2-oxoglutarate adduct) | | Descriptor: | 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase, 4-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-5-[2-[oxidanyl(phosphonooxy)phosphoryl]oxyethyl]-1,3-thiazol-3 -ium-2-yl]-4-oxidanyl-butanoic acid, ACETATE ION, ... | | Authors: | Johnston, J.M, Jirgis, E.N.M, Bashiri, G, Bulloch, E.M.M, Baker, E.N. | | Deposit date: | 2015-11-17 | | Release date: | 2016-06-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Views along the Mycobacterium tuberculosis MenD Reaction Pathway Illuminate Key Aspects of Thiamin Diphosphate-Dependent Enzyme Mechanisms.
Structure, 24, 2016
|
|
7G04
 
 | | Crystal Structure of human FABP5 in complex with 7-(4-chlorophenyl)-1,2,3,4-tetrahydronaphthalene-1-carboxylic acid, i.e. SMILES c12c(ccc(c2)c2ccc(cc2)Cl)CCC[C@H]1C(=O)O with IC50=3.3 microM | | Descriptor: | (1R)-7-(4-chlorophenyl)-1,2,3,4-tetrahydronaphthalene-1-carboxylic acid, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Canesso, R, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
6LMR
 
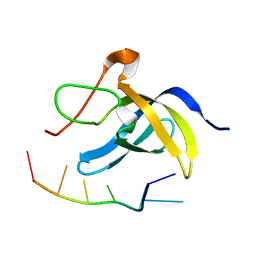 | | Solution structure of cold shock domain and ssDNA complex | | Descriptor: | DNA (5'-D(P*AP*AP*CP*AP*CP*CP*T)-3'), Y-box-binding protein 1 | | Authors: | Fan, J, Yang, D. | | Deposit date: | 2019-12-26 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of DNA binding to human YB-1 cold shock domain regulated by phosphorylation.
Nucleic Acids Res., 48, 2020
|
|
4JVR
 
 | | Co-crystal structure of MDM2 with inhibitor (2'S,3R,4'S,5'R)-N-(2-aminoethyl)-6-chloro-4'-(3-chloro-2-fluorophenyl)-2'-(2,2-dimethylpropyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxamide | | Descriptor: | (2'S,3R,4'S,5'R)-N-(2-aminoethyl)-6-chloro-4'-(3-chloro-2-fluorophenyl)-2'-(2,2-dimethylpropyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxamide, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Huang, X, Gonzalez-Lopez de Turiso, F, Sun, D, Yosup, R, Bartberger, M.D, Beck, H.P, Cannon, J, Shaffer, P, Oliner, J.D, Olson, S.H, Medina, J.C. | | Deposit date: | 2013-03-26 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational Design and Binding Mode Duality of MDM2-p53 Inhibitors.
J.Med.Chem., 56, 2013
|
|
6RRW
 
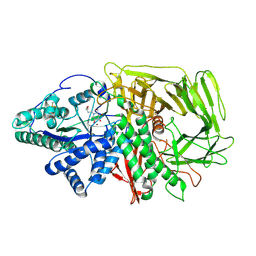 | | GOLGI ALPHA-MANNOSIDASE II in complex with (2R,3R,4R,5S)-1-(5-{[4-(3,4-Dihydro-2H-1,5-benzodioxepin-7-yl)benzyl]oxy}pentyl)-2-(hydroxymethyl)-3,4,5-piperidinetriol | | Descriptor: | (2~{R},3~{R},4~{R},5~{S})-1-[5-[[4-(3,4-dihydro-2~{H}-1,5-benzodioxepin-7-yl)phenyl]methoxy]pentyl]-2-(hydroxymethyl)piperidine-3,4,5-triol, Alpha-mannosidase 2, ZINC ION | | Authors: | Armstrong, Z, Lahav, D, Johnson, R, Kuo, C.L, Beenakker, T.J.M, de Boer, C, Wong, C.S, van Rijssel, E.R, Debets, M, Geurink, P.P, Ovaa, H, van der Stelt, M, Codee, J.D.C, Aerts, J.M.F.G, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2019-05-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Manno- epi -cyclophellitols Enable Activity-Based Protein Profiling of Human alpha-Mannosidases and Discovery of New Golgi Mannosidase II Inhibitors.
J.Am.Chem.Soc., 142, 2020
|
|
4BDC
 
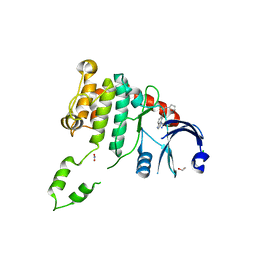 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, N-(furan-2-ylmethyl)quinoxaline-6-carboxamide, ... | | Authors: | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
5ICP
 
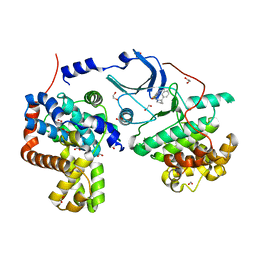 | | CDK8-CYCC IN COMPLEX WITH [(S)-2-(4-Chloro-phenyl)-pyrrolidin-1-yl]-(5-methyl-imidazo[5,1-b][1,3,4]thiadiazol-2-yl)-methanone | | Descriptor: | 1,2-ETHANEDIOL, Cyclin-C, Cyclin-dependent kinase 8, ... | | Authors: | Musil, D, Blagg, J, Mallinger, A, Czodrowski, P, Schiemann, K. | | Deposit date: | 2016-02-23 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure-Based Optimization of Potent, Selective, and Orally Bioavailable CDK8 Inhibitors Discovered by High-Throughput Screening.
J. Med. Chem., 59, 2016
|
|
4O8K
 
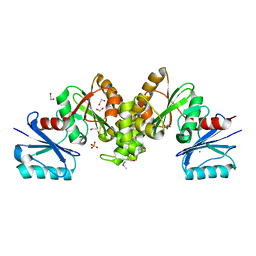 | |
5ETN
 
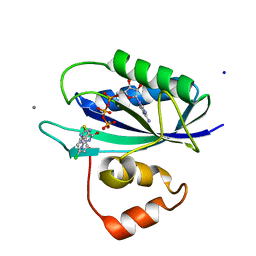 | | E. coli 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase complexed with AMPCPP and inhibitor at 1.40 angstrom resolution | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 2-azanyl-8-[(4-chlorophenyl)methylsulfanyl]-1,9-dihydropurin-6-one, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Dennis, M.L, Peat, T.S, Swarbrick, J.D. | | Deposit date: | 2015-11-17 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis for the Selective Binding of Inhibitors to 6-Hydroxymethyl-7,8-dihydropterin Pyrophosphokinase from Staphylococcus aureus and Escherichia coli.
J.Med.Chem., 59, 2016
|
|
6C0S
 
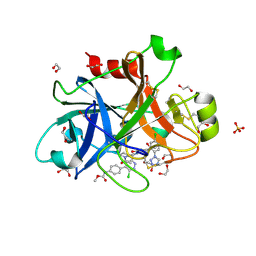 | |
5EQP
 
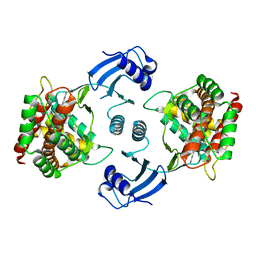 | | Crystal structure of choline kinase alpha-1 bound by 6-[(4-methyl-1,4-diazepan-1-yl)methyl]quinoline (compound 37) | | Descriptor: | 6-[(4-methyl-1,4-diazepan-1-yl)methyl]quinoline, Choline kinase alpha | | Authors: | Zhou, T, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2015-11-13 | | Release date: | 2016-01-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Novel Small Molecule Inhibitors of Choline Kinase Identified by Fragment-Based Drug Discovery.
J.Med.Chem., 59, 2016
|
|
5ETO
 
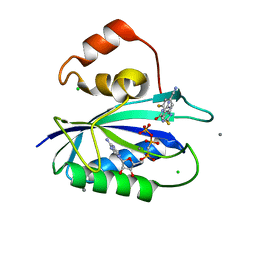 | | E. coli 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase complexed with AMPCPP and inhibitor at 1.07 angstrom resolution | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 4-[(2-azanyl-6-oxidanylidene-1,9-dihydropurin-8-yl)sulfanylmethyl]-3-fluoranyl-benzenecarbonitrile, CALCIUM ION, ... | | Authors: | Dennis, M.L, Peat, T.S, Swarbrick, J.D. | | Deposit date: | 2015-11-17 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Structural Basis for the Selective Binding of Inhibitors to 6-Hydroxymethyl-7,8-dihydropterin Pyrophosphokinase from Staphylococcus aureus and Escherichia coli.
J.Med.Chem., 59, 2016
|
|
3JVE
 
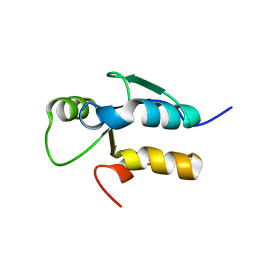 | |
3K2K
 
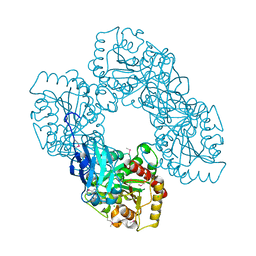 | |
5V7I
 
 | | Crystal structure of homo sapiens serine hydroxymethyltransferase 2 (mitochondrial) (SHMT2), in complex with glycine, PLP and folate-competitive pyrazolopyran inhibitor: 6-amino-4-isopropyl-3-methyl-4-(3-(pyrrolidin-1-yl)-5-(trifluoromethyl)phenyl)-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile | | Descriptor: | (4R)-6-amino-3-methyl-4-(propan-2-yl)-4-[3-(pyrrolidin-1-yl)-5-(trifluoromethyl)phenyl]-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Ducker, G.S, Ghergurovich, J.M, Mainolfi, N, Suri, V, Jeong, S, Friedman, A, Manfredi, M, Kim, H, Rabinowitz, J.D. | | Deposit date: | 2017-03-20 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Human SHMT inhibitors reveal defective glycine import as a targetable metabolic vulnerability of diffuse large B-cell lymphoma.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5ETY
 
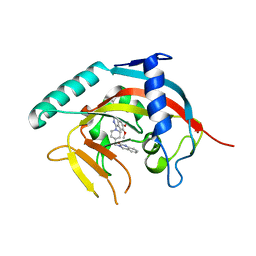 | | Crystal Structure of human Tankyrase-1 bound to K-756 | | Descriptor: | 3-[[1-(6,7-dimethoxyquinazolin-4-yl)piperidin-4-yl]methyl]-1,4-dihydroquinazolin-2-one, Tankyrase-1, ZINC ION | | Authors: | Takahashi, Y, Miyagi, H, Suzuki, M, Saito, J. | | Deposit date: | 2015-11-18 | | Release date: | 2016-06-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Discovery and Characterization of K-756, a Novel Wnt/ beta-Catenin Pathway Inhibitor Targeting Tankyrase
Mol.Cancer Ther., 15, 2016
|
|
6UVE
 
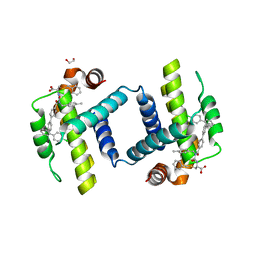 | | Crystal structure of BCL-XL bound to compound 7: (R)-3-(Benzylthio)-2-(3-(4-chloro-[1,1':2',1'':3'',1'''-quaterphenyl]-4'''-carbonyl)-3-(4-methylbenzyl)ureido)propanoic acid | | Descriptor: | (R)-3-(Benzylthio)-2-(3-(4-chloro-[1,1':2',1'':3'',1'''-quaterphenyl]-4'''-carbonyl)-3-(4-methylbenzyl)ureido)propanoic acid, 1,2-ETHANEDIOL, Bcl-2-like protein 1 | | Authors: | Roy, M.J, Lessene, G, Czabotar, P.E. | | Deposit date: | 2019-11-02 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structure-Guided Development of Potent Benzoylurea Inhibitors of BCL-X L and BCL-2.
J.Med.Chem., 64, 2021
|
|
4BDE
 
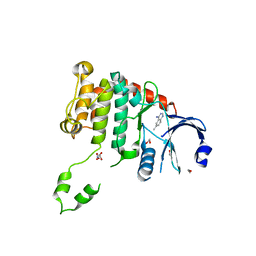 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | Descriptor: | 1,2-ETHANEDIOL, 6-METHYLQUINAZOLIN-4-AMINE, NITRATE ION, ... | | Authors: | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
6UVH
 
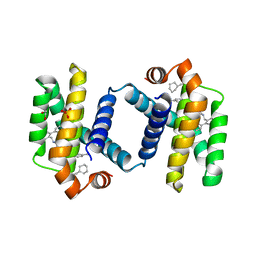 | | Crystal structure of BCL-XL bound to compound 15: (R)-2-(3-(2-((4'-Chloro-[1,1'-biphenyl]-2-yl)methyl)-1,2,3,4-tetrahydroisoquinoline-6-carbonyl)-3-(4-methylbenzyl)ureido)-3-((cyclohexylmethyl)sulfonyl)propanoic acid | | Descriptor: | (R)-2-(3-(2-((4'-Chloro-[1,1'-biphenyl]-2-yl)methyl)-1,2,3,4-tetrahydroisoquinoline-6-carbonyl)-3-(4-methylbenzyl)ureido)-3-((cyclohexylmethyl)sulfonyl)propanoic acid, 1,2-ETHANEDIOL, Bcl-2-like protein 1, ... | | Authors: | Roy, M.J, Lessene, G, Czabotar, P.E. | | Deposit date: | 2019-11-02 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure-Guided Development of Potent Benzoylurea Inhibitors of BCL-X L and BCL-2.
J.Med.Chem., 64, 2021
|
|
6UVG
 
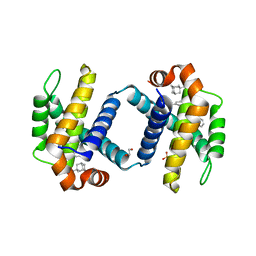 | | Crystal structure of BCL-XL bound to compound 13: (R)-2-(3-([1,1'-Biphenyl]-4-carbonyl)-3-(4-methylbenzyl)ureido)-3-(((3R,5R,7R)-adamantan-1-ylmethyl)sulfonyl)propanoic acid | | Descriptor: | (R)-2-(3-([1,1'-Biphenyl]-4-carbonyl)-3-(4-methylbenzyl)ureido)-3-(((3R,5R,7R)-adamantan-1-ylmethyl)sulfonyl)propanoic acid, 1,2-ETHANEDIOL, Bcl-2-like protein 1, ... | | Authors: | Roy, M.J, Lessene, G, Czabotar, P.E. | | Deposit date: | 2019-11-02 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Development of Potent Benzoylurea Inhibitors of BCL-X L and BCL-2.
J.Med.Chem., 64, 2021
|
|
6S8X
 
 | | Crystal structure of the Rab-binding domain of FIP2 | | Descriptor: | HEXANE-1,6-DIOL, Rab11 family-interacting protein 2 | | Authors: | Kearney, A.M, Khan, A.R. | | Deposit date: | 2019-07-10 | | Release date: | 2020-08-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of the Rab-binding domain of Rab11 family-interacting protein 2.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
5ZLM
 
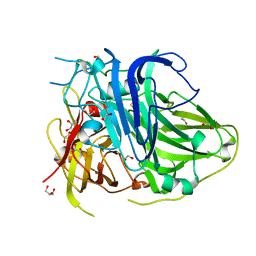 | | Mutation in the trinuclear site of CotA-laccase: H491C mutant, PH 8.0 | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, GLYCEROL, ... | | Authors: | Xie, T, Liu, Z.C, Wang, G.G. | | Deposit date: | 2018-03-28 | | Release date: | 2018-05-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insight into the Allosteric Coupling of Cu1 Site and Trinuclear Cu Cluster in CotA Laccase.
Chembiochem, 19, 2018
|
|
5EU9
 
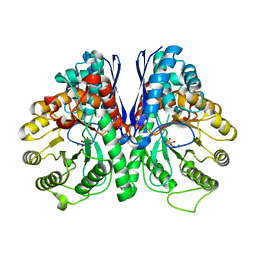 | | Structure of Human Enolase 2 in complex with ((3S,5S)-1,5-dihydroxy-3-methyl-2-oxopyrrolidin-3-yl)phosphonic acid | | Descriptor: | ((3S,5S)-1,5-dihydroxy-3-methyl-2-oxopyrrolidin-3-yl)phosphonic acid, Gamma-enolase, MAGNESIUM ION, ... | | Authors: | Leonard, P.G, Muller, F.L. | | Deposit date: | 2015-11-18 | | Release date: | 2016-11-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | SF2312, a natural phosphonate inhibitor of Enolase
To be Published
|
|
5V09
 
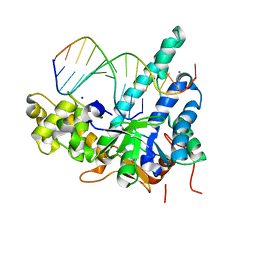 | | Crystal structure of human exonuclease 1 Exo1 (D225A) in complex with 5' recessed-end DNA (rVII) | | Descriptor: | DNA (5'-D(*CP*GP*CP*TP*AP*GP*TP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(P*AP*CP*GP*AP*CP*TP*AP*GP*CP*G)-3'), Exonuclease 1, ... | | Authors: | Shi, Y, Beese, L.S. | | Deposit date: | 2017-02-28 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Interplay of catalysis, fidelity, threading, and processivity in the exo- and endonucleolytic reactions of human exonuclease I.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
