7G1U
 
 | | Crystal Structure of human FABP4 in complex with 2-[[5-chloro-2-(3-methyl-1,2,4-oxadiazol-5-yl)phenyl]carbamoyl]cyclopentene-1-carboxylic acid, i.e. SMILES N(c1cc(ccc1C1=NC(=NO1)C)Cl)C(=O)C1=C(CCC1)C(=O)O with IC50=0.195 microM | | Descriptor: | 2-{[(2M)-5-chloro-2-(3-methyl-1,2,4-oxadiazol-5-yl)phenyl]carbamoyl}cyclopent-1-ene-1-carboxylic acid, DIMETHYL SULFOXIDE, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Richter, H, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
5EL1
 
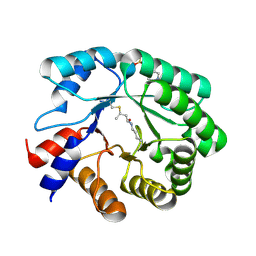 | |
5HBE
 
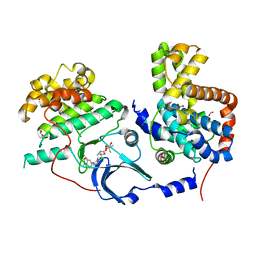 | | CDK8-CYCC IN COMPLEX WITH 8-[3-Chloro-5-(1-methyl-2,2-dioxo-2, 3-dihydro-1H-2l6-benzo[c]isothiazol-5-yl)-pyridin- 4-yl]-1-oxa-3,8-diaza-spiro[4.5]decan-2-one | | Descriptor: | 1,2-ETHANEDIOL, 8-[3-chloranyl-5-[1-methyl-2,2-bis(oxidanylidene)-3~{H}-2,1-benzothiazol-5-yl]pyridin-4-yl]-1-oxa-3,8-diazaspiro[4.5]decan-2-one, Cyclin-C, ... | | Authors: | Musil, D, Blagg, J, Mallinger, A. | | Deposit date: | 2015-12-31 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Discovery of Potent, Selective, and Orally Bioavailable Small-Molecule Modulators of the Mediator Complex-Associated Kinases CDK8 and CDK19.
J.Med.Chem., 59, 2016
|
|
2OGH
 
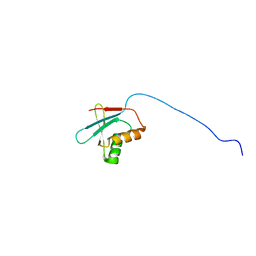 | | Solution structure of yeast eIF1 | | Descriptor: | Eukaryotic translation initiation factor eIF-1 | | Authors: | Reibarkh, M, del Rio, F, Yamamoto, Y, Asano, K, Wagner, G. | | Deposit date: | 2007-01-05 | | Release date: | 2007-11-20 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Eukaryotic Initiation Factor (eIF) 1 Carries Two Distinct eIF5-binding Faces Important for Multifactor Assembly and AUG Selection.
J.Biol.Chem., 283, 2008
|
|
7G0B
 
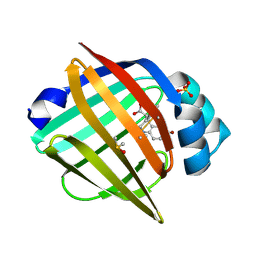 | | Crystal Structure of human FABP5 in complex with 7-bromo-1-methyl-5-phenyl-2,3,4,5-tetrahydro-1-benzazepine-4-carboxylic acid, i.e. SMILES [C@@H]1([C@@H](CCN(c2c1cc(cc2)Br)C)C(=O)O)c1ccccc1 with IC50=2.3 microM | | Descriptor: | (4S,5S)-7-bromo-1-methyl-5-phenyl-2,3,4,5-tetrahydro-1H-1-benzazepine-4-carboxylic acid, DIMETHYL SULFOXIDE, Fatty acid-binding protein 5, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Ceccarelli-Simona, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
6MAW
 
 | | F9 Pilus Adhesin FmlH Lectin Domain from E. coli UTI89 in Complex with Galactoside N-[(2S,3R,4R,5R,6R)-4,5-dihydroxy-6-(hydroxymethyl)-2-{[S-methyl-6-(trifluoromethyl)-[1,1'-biphenyl]-3'-yl]oxy}oxan-3-yl]acetamide | | Descriptor: | Fimbrial adhesin FmlD, N-[2'-{[2-(acetylamino)-2-deoxy-beta-D-galactopyranosyl]oxy}-6'-(trifluoromethyl)[1,1'-biphenyl]-3-yl]methanesulfonamide | | Authors: | Klein, R.D, Hultgren, S.J. | | Deposit date: | 2018-08-28 | | Release date: | 2019-09-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Biphenyl Gal and GalNAc FmlH Lectin Antagonists of Uropathogenic E. coli (UPEC): Optimization through Iterative Rational Drug Design.
J.Med.Chem., 62, 2019
|
|
5EQE
 
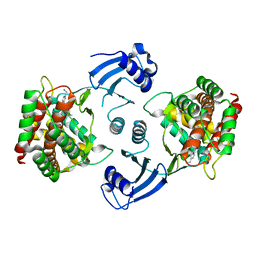 | | Crystal structure of choline kinase alpha-1 bound by [4-[(4-methyl-1,4-diazepan-1-yl)methyl]phenyl]methanamine (compound 11) | | Descriptor: | Choline kinase alpha, [4-[(4-methyl-1,4-diazepan-1-yl)methyl]phenyl]methanamine | | Authors: | Zhou, T, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2015-11-12 | | Release date: | 2016-01-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Novel Small Molecule Inhibitors of Choline Kinase Identified by Fragment-Based Drug Discovery.
J.Med.Chem., 59, 2016
|
|
7G1J
 
 | | Crystal Structure of human FABP4 in complex with 2-[[3-(3-cyclopropyl-1,2,4-oxadiazol-5-yl)-4,5-dimethylthiophen-2-yl]carbamoyl]cyclohexene-1-carboxylic acid, i.e. SMILES C1(=C(CCCC1)C(=O)NC1=C(C(=C(C)S1)C)C1=NC(=NO1)C1CC1)C(=O)O with IC50=0.0236791 microM | | Descriptor: | 2-{[(3M)-3-(3-cyclopropyl-1,2,4-oxadiazol-5-yl)-4,5-dimethylthiophen-2-yl]carbamoyl}cyclohex-1-ene-1-carboxylic acid, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Richter, H, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
7FWI
 
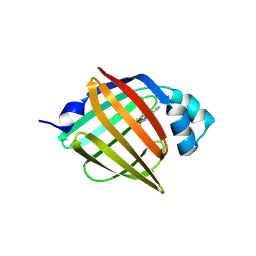 | | Crystal Structure of human FABP5 in complex with 2-(indole-1-carbonylamino)benzoic acid, i.e. SMILES c12N(C(=O)Nc3c(cccc3)C(=O)O)C=Cc1cccc2 with IC50=18.1696 microM | | Descriptor: | 2-[(2,3-dihydro-1H-indole-1-carbonyl)amino]benzoic acid, CHLORIDE ION, Fatty acid-binding protein 5 | | Authors: | Ehler, A, Benz, J, Obst, U, Ning, R, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
7FX5
 
 | | Crystal Structure of human FABP4 in complex with 5-[[3-(3-cyclopropyl-1,2,4-oxadiazol-5-yl)-6,6-difluoro-5,7-dihydro-4H-1-benzothiophen-2-yl]carbamoyl]-3,6-dihydro-2H-pyran-4-carboxylic acid, i.e. SMILES C1C(CC2=C(C1)C(=C(S2)NC(=O)C1=C(CCOC1)C(=O)O)C1=NC(=NO1)C1CC1)(F)F with IC50=0.182863 microM | | Descriptor: | 5-{[(3M)-3-(3-cyclopropyl-1,2,4-oxadiazol-5-yl)-6,6-difluoro-4,5,6,7-tetrahydro-1-benzothiophen-2-yl]carbamoyl}-3,6-dihydro-2H-pyran-4-carboxylic acid, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Richter, H, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
5UK8
 
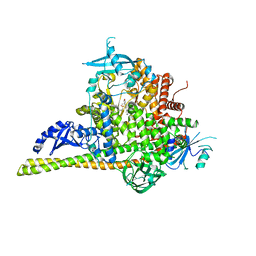 | | The co-structure of (R)-4-(6-(1-(cyclopropylsulfonyl)cyclopropyl)-2-(1H-indol-4-yl)pyrimidin-4-yl)-3-methylmorpholine and a rationally designed PI3K-alpha mutant that mimics ATR | | Descriptor: | (R)-4-(6-(1-(cyclopropylsulfonyl)cyclopropyl)-2-(1H-indol-4-yl)pyrimidin-4-yl)-3-methylmorpholine, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Knapp, M.S, Mamo, M, Elling, R.A. | | Deposit date: | 2017-01-20 | | Release date: | 2017-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rationally Designed PI3K alpha Mutants to Mimic ATR and Their Use to Understand Binding Specificity of ATR Inhibitors.
J. Mol. Biol., 429, 2017
|
|
7XGG
 
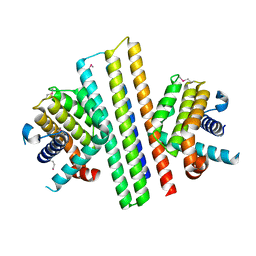 | |
7FZI
 
 | | Crystal Structure of human FABP4 in complex with 2-[[2-(3-cyclopropyl-1,2,4-oxadiazol-5-yl)phenyl]carbamoyl]cyclopentene-1-carboxylic acid, i.e. SMILES N(c1ccccc1C1=NC(=NO1)C1CC1)C(=O)C1=C(CCC1)C(=O)O with IC50=0.0623077 microM | | Descriptor: | 2-{[(2M)-2-(3-cyclopropyl-1,2,4-oxadiazol-5-yl)phenyl]carbamoyl}cyclopent-1-ene-1-carboxylic acid, DIMETHYL SULFOXIDE, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Richter, H, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
5ELJ
 
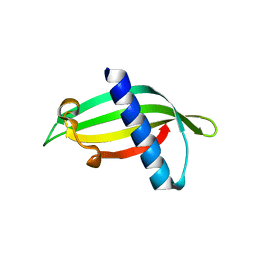 | | Isoform-specific inhibition of SUMO-dependent protein-protein interactions | | Descriptor: | SUMO-Affirmer-S2D5, Small ubiquitin-related modifier 1 | | Authors: | Hughes, D.J, Tiede, C, Hall, N, Tang, A.A.S, Trinh, C.H, Zajac, K, Mandal, U, Howell, G, Edwards, T.A, McPherson, M.J, Tomlinson, D.C, Whitehouse, A. | | Deposit date: | 2015-11-04 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.983 Å) | | Cite: | Generation of specific inhibitors of SUMO-1- and SUMO-2/3-mediated protein-protein interactions using Affimer (Adhiron) technology.
Sci Signal, 10, 2017
|
|
7FZW
 
 | | Crystal Structure of human FABP4 in complex with 2-[[5-bromo-2-(3-cyclopropyl-1,2,4-oxadiazol-5-yl)phenyl]carbamoyl]cyclopentene-1-carboxylic acid, i.e. SMILES C1(=C(CCC1)C(=O)O)C(=O)Nc1cc(ccc1C1=NC(=NO1)C1CC1)Br with IC50=0.333165 microM | | Descriptor: | 2-{[(2M)-5-bromo-2-(3-cyclopropyl-1,2,4-oxadiazol-5-yl)phenyl]carbamoyl}cyclopent-1-ene-1-carboxylic acid, BROMIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Richter, H, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
6LRE
 
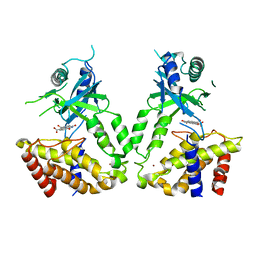 | | Human cGAS catalytic domain bound with compound 3 | | Descriptor: | 1,3-bis(oxidanylidene)benzo[de]isoquinoline-6,7-dicarboxylic acid, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Zhao, W.F, Xiong, M.Y, Yuan, X.J, Sun, H.B, Xu, Y.C. | | Deposit date: | 2020-01-16 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | In Silico Screening-Based Discovery of Novel Inhibitors of Human Cyclic GMP-AMP Synthase: A Cross-Validation Study of Molecular Docking and Experimental Testing.
J.Chem.Inf.Model., 60, 2020
|
|
3ARA
 
 | | Discovery of Novel Uracil Derivatives as Potent Human dUTPase Inhibitors | | Descriptor: | 1-[3-({(2R)-2-[hydroxy(diphenyl)methyl]pyrrolidin-1-yl}sulfonyl)propyl]pyrimidine-2,4(1H,3H)-dione, Deoxyuridine 5'-triphosphate nucleotidohydrolase, MAGNESIUM ION | | Authors: | Chong, K.T, Miyakoshi, H, Miyahara, S, Fukuoka, M. | | Deposit date: | 2010-11-25 | | Release date: | 2010-12-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Synthesis and discovery of N-carbonylpyrrolidine- or N-sulfonylpyrrolidine-containing uracil derivatives as potent human deoxyuridine triphosphatase inhibitors
J.Med.Chem., 55, 2012
|
|
5UWX
 
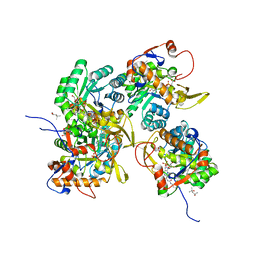 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and P176 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, ... | | Authors: | Maltseva, N, Kim, Y, Mulligan, R, Makowska-Grzyska, M, Gu, M, Gollapalli, D.R, Hedstrom, L, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-21 | | Release date: | 2017-03-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from
Clostridium perfringens
Complexed with IMP and P176
To Be Published
|
|
6CQ5
 
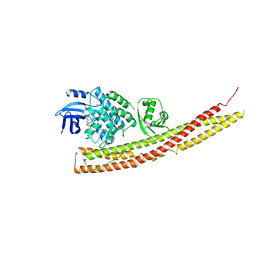 | | TBK1 in Complex with Sulfone Analog of Amlexanox | | Descriptor: | 2-amino-7-(1,1-dioxo-1lambda~6~-thian-4-yl)-5-oxo-5H-[1]benzopyrano[2,3-b]pyridine-3-carboxylic acid, Serine/threonine-protein kinase TBK1 | | Authors: | Beyett, T.S, Tesmer, J.J.G. | | Deposit date: | 2018-03-14 | | Release date: | 2018-12-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.354 Å) | | Cite: | Design, synthesis, and biological activity of substituted 2-amino-5-oxo-5H-chromeno[2,3-b]pyridine-3-carboxylic acid derivatives as inhibitors of the inflammatory kinases TBK1 and IKK epsilon for the treatment of obesity.
Bioorg. Med. Chem., 26, 2018
|
|
7FZL
 
 | | Crystal Structure of human FABP4 in complex with 3-(3-chlorophenyl)-4-prop-2-enyl-1H-1,2,4-triazole-5-thione, i.e. SMILES N1(C(=NNC1=S)c1cc(Cl)ccc1)CC=C with IC50=1.4 microM | | Descriptor: | (5P)-5-(3-chlorophenyl)-4-(prop-2-en-1-yl)-2,4-dihydro-3H-1,2,4-triazole-3-thione, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Brunner, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
4CXQ
 
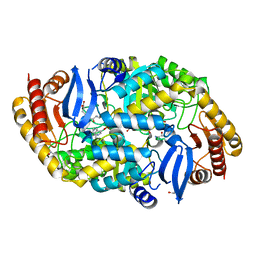 | | Mycobaterium tuberculosis transaminase BioA complexed with substrate KAPA | | Descriptor: | 1,2-ETHANEDIOL, 7-KETO-8-AMINOPELARGONIC ACID, ADENOSYLMETHIONINE-8-AMINO-7-OXONONANOATE AMINOTRANSFERASE, ... | | Authors: | Dai, R, Wilson, D.J, Geders, T.W, Aldrich, C.C, Finzel, B.C. | | Deposit date: | 2014-04-08 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibition of Mycobacterium Tuberculosis Transaminase Bioa by Aryl Hydrazines and Hydrazides.
Chembiochem, 15, 2014
|
|
7B2H
 
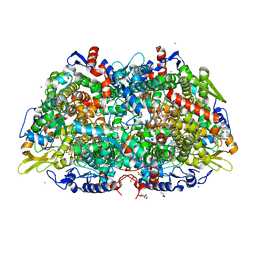 | | Crystal structure of the methyl-coenzyme M reductase from Methanothermobacter Marburgensis derivatized with xenon | | Descriptor: | 1,2-ETHANEDIOL, 1-THIOETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Wagner, T, Lemaire, O.N, Engilberge, S. | | Deposit date: | 2020-11-27 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structure of a key enzyme for anaerobic ethane activation.
Science, 373, 2021
|
|
7FXZ
 
 | | Crystal Structure of human FABP4 in complex with 4-hexylsulfanyl-1-(3-pyridylmethyl)-6-sulfanyl-1,3,5-triazin-2-one | | Descriptor: | 4-hexylsulfanyl-1-(pyridin-3-ylmethyl)-6-sulfanyl-1$l^{4},3,5-triazacyclohexa-1,3,5-trien-2-ol, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
1OSW
 
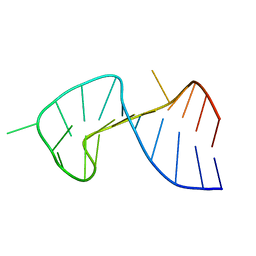 | | The Stem of SL1 RNA in HIV-1: Structure and Nucleocapsid Protein Binding for a 1X3 Internal Loop | | Descriptor: | 5'-R(*GP*GP*AP*GP*GP*CP*GP*CP*UP*AP*CP*GP*GP*CP*GP*AP*GP*GP*CP*UP*CP*CP*A)-3' | | Authors: | Yuan, Y, Kerwood, D.J, Paoletti, A.C, Shubsda, M.F, Borer, P.N. | | Deposit date: | 2003-03-20 | | Release date: | 2003-05-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Stem of SL1 RNA in HIV-1: Structure and Nucleocapsid Protein Binding for a 1X3 Internal Loop
Biochemistry, 42, 2003
|
|
3AXB
 
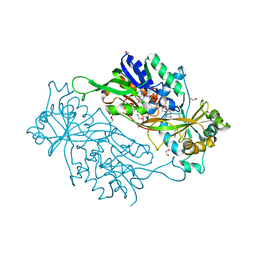 | | Structure of a dye-linked L-proline dehydrogenase from the aerobic hyperthermophilic archaeon, Aeropyrum pernix | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, PROLINE, ... | | Authors: | Sakuraba, H, Ohshima, T, Satomura, T, Yoneda, K, Hara, Y. | | Deposit date: | 2011-04-01 | | Release date: | 2012-04-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of Novel Dye-linked L-Proline Dehydrogenase from Hyperthermophilic Archaeon Aeropyrum pernix
J.Biol.Chem., 287, 2012
|
|
