5AH9
 
 | | Disubstituted bis-THF moieties as new P2 ligands in non-peptidal HIV- 1 Protease Inhibitors (II) | | Descriptor: | (3R,3aS,4R,6aR)-4-(2-methoxyethoxy)hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{[(4-aminophenyl)sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, PROTEASE | | Authors: | Hohlfeld, K, Wegner, J.K, Kesteleyn, B, Linclau, B, Enstrom, O, Unge, J. | | Deposit date: | 2015-02-05 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Disubstituted Bis-Thf Moieties as New P2 Ligands in Non-Peptidal HIV-1 Protease Inhibitors (II).
J.Med.Chem., 58, 2015
|
|
5BN2
 
 | | Room Temperature Structure of Pichia pastoris aquaporin at 1.3 A | | Descriptor: | AQY1 protein, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Fischer, G, Kosinska Eriksson, U, Hedfalk, K, Neutze, R. | | Deposit date: | 2015-05-25 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Room Temperature Structure of Pichia pastoris aquaporin at 1.3 A
To Be Published
|
|
4ZZW
 
 | | Geotrichum candidum Cel7A structure complex with cellobiose at 1.5A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE I, ... | | Authors: | Borisova, A.S, Stahlberg, J. | | Deposit date: | 2015-04-14 | | Release date: | 2015-09-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Sequencing, Biochemical Characterization, Crystal Structure and Molecular Dynamics of Cellobiohydrolase Cel7A from Geotrichum Candidum 3C.
FEBS J., 282, 2015
|
|
5ACI
 
 | | X-ray Structure of LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Frandsen, K.E.H, Poulsen, J.N, Tovborg, M, Johanson, K.S, Lo Leggio, L. | | Deposit date: | 2015-08-17 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The molecular basis of polysaccharide cleavage by lytic polysaccharide monooxygenases.
Nat. Chem. Biol., 12, 2016
|
|
4ZZQ
 
 | | Dictyostelium discoideum cellobiohydrolase Cel7A apo structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLULOSE 1,4-BETA-CELLOBIOSIDASE, DI(HYDROXYETHYL)ETHER | | Authors: | Momeni, M.H, Hobdey, S.E, Knott, B, Beckham, G.T, Stahlberg, J. | | Deposit date: | 2015-04-13 | | Release date: | 2016-04-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical and Structural Characterization of Two Dictyostelium Cellobiohydrolases from the Amoebozoa Kingdom Reveal a High Conservation between Distant Phylogenetic Trees of Life.
Appl.Environ.Microbiol., 82, 2016
|
|
4ZZU
 
 | | Geotrichum candidum Cel7A structure complex with thio-linked cellotetraose at 1.4A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE I, ... | | Authors: | Borisova, A.S, Stahlberg, J. | | Deposit date: | 2015-04-14 | | Release date: | 2015-09-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Sequencing, Biochemical Characterization, Crystal Structure and Molecular Dynamics of Cellobiohydrolase Cel7A from Geotrichum Candidum 3C.
FEBS J., 282, 2015
|
|
5B0W
 
 | |
5ACJ
 
 | | X-ray Structure of LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Frandsen, K.E.H, Poulsen, J.N, Tovborg, M, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2015-08-17 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The molecular basis of polysaccharide cleavage by lytic polysaccharide monooxygenases.
Nat. Chem. Biol., 12, 2016
|
|
5A0Z
 
 | |
5AMD
 
 | |
6OHD
 
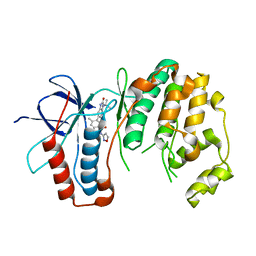 | | P38 in complex with T-3220137 | | Descriptor: | 3-(3-tert-butyl-2-oxo-2,3-dihydro-1H-imidazo[4,5-b]pyridin-6-yl)-4-methyl-N-(1,2-oxazol-3-yl)benzamide, Mitogen-activated protein kinase 14 | | Authors: | Lane, W, Saikatendu, K. | | Deposit date: | 2019-04-05 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Design, Synthesis, and Biological Evaluation of Imidazo[4,5-b]Pyridin-2-one-Based p38 MAP Kinase Inhibitors: Part 2.
Chemmedchem, 14, 2019
|
|
5AHC
 
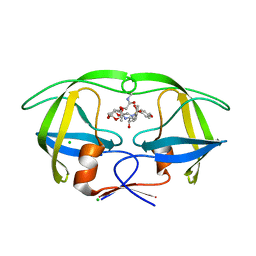 | | Disubstituted bis-THF moieties as new P2 ligands in non-peptidal HIV- 1 Protease Inhibitors (II) | | Descriptor: | (3R,3aS,4R,6aR)-4-[2-(methylamino)-2-oxoethoxy]hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, PROTEASE | | Authors: | Hohlfeld, K, Wegner, J.K, Kesteleyn, B, Linclau, B, Unge, J. | | Deposit date: | 2015-02-05 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Disubstituted Bis-Thf Moieties as New P2 Ligands in Non-Peptidal HIV-1 Protease Inhibitors (II).
J.Med.Chem., 58, 2015
|
|
5A4J
 
 | | Crystal structure of FTHFS1 from T.acetoxydans Re1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, D(-)-TARTARIC ACID, ... | | Authors: | Bergdahl, R, Jacobson, F, Muller, B, Mikkelsen, N, Schurer, A, Sandgren, M. | | Deposit date: | 2015-06-10 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Characterization, Crystallization and Three- Dimensional Structures of Formyltetrahydrofolate Synthetase (Fthfs) from the Syntrophic Acetate Oxidising Bacterium Tepidanaerobacter Acetatoxydans Re1
To be Published
|
|
5A7C
 
 | | Crystal structure of the second bromodomain of human BRD3 in complex with compound | | Descriptor: | 1,2-ETHANEDIOL, BROMODOMAIN-CONTAINING PROTEIN 3, N-(6-ACETAMIDOHEXYL)ACETAMIDE | | Authors: | Welin, M, Kimbung, R, Diehl, C, Hakansson, M, Logan, D.T, Walse, B. | | Deposit date: | 2015-07-03 | | Release date: | 2016-03-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cancer Differentiating Agent Hexamethylene Bisacetamide Inhibits Bet Bromodomain Proteins.
Cancer Res., 76, 2016
|
|
5AML
 
 | |
5AMG
 
 | |
8S79
 
 | | Lotus japonicus NFR5 intracellular domain in complex with Nanobody 200 | | Descriptor: | ACETATE ION, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hansen, S.B, Gysel, K, Andersen, K.R. | | Deposit date: | 2024-02-29 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A conserved juxtamembrane motif in plant NFR5 receptors is essential for root nodule symbiosis.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8RNE
 
 | | HLA-E*01:03 in complex with SARS-CoV-2 Nsp13 peptide, VMPLSAPTL | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, E alpha chain variant, ... | | Authors: | Sun, R, Achour, A, Sala, B.M, Sandalova, T. | | Deposit date: | 2024-01-09 | | Release date: | 2024-12-04 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Emerging mutation in SARS-CoV-2 facilitates escape from NK cell recognition and associates with enhanced viral fitness.
Plos Pathog., 20, 2024
|
|
8RNF
 
 | | HLA-E*01:03 in complex with SARS-CoV-2 Omicron Nsp13 peptide, VIPLSAPTL | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, E alpha chain variant, ... | | Authors: | Sun, R, Achour, A, Sala, B.M, Sandalova, T. | | Deposit date: | 2024-01-09 | | Release date: | 2024-12-04 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.872 Å) | | Cite: | Emerging mutation in SARS-CoV-2 facilitates escape from NK cell recognition and associates with enhanced viral fitness.
Plos Pathog., 20, 2024
|
|
8S6Z
 
 | | CD28 in complex with the antibody Fab fragment AI3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Majocchi, S, Giovanni, M, Malinge, P, Fischer, N, Svensson, L.A, Kelpsas, V, Rose, N.C. | | Deposit date: | 2024-02-28 | | Release date: | 2024-12-18 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | NI-3201 Is a Bispecific Antibody Mediating PD-L1-Dependent CD28 Co-stimulation on T Cells for Enhanced Tumor Control.
Cancer Immunol Res, 13, 2025
|
|
1LDO
 
 | | avidin-norbioitn complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NORBIOTIN, avidin | | Authors: | Pazy, Y, Kulik, T, Bayer, E.A, Wilchek, M, Livnah, O. | | Deposit date: | 2002-04-09 | | Release date: | 2002-11-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ligand exchange between proteins: exchange of biotin and biotin derivatives between avidin and streptavidin
J.Biol.Chem., 277, 2002
|
|
8S1Z
 
 | | Crystal structure of glycosylated human primary amine oxidase AOC3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guedez, G, Alix, M, Salminen, T.A. | | Deposit date: | 2024-02-16 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural flexibility of domains in human AOC3
To Be Published
|
|
8S60
 
 | | Tankyrase 2 in complex with a quinazolin-4-one inhibitor | | Descriptor: | 8-nitro-2-[4-(trifluoromethyl)phenyl]-3H-quinazolin-4-one, GLYCEROL, Poly [ADP-ribose] polymerase tankyrase-2, ... | | Authors: | Bosetti, C, Paakkonen, J, Lehtio, L. | | Deposit date: | 2024-02-26 | | Release date: | 2025-03-12 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Substitutions at the C-8 position of quinazolin-4-ones improve the potency of nicotinamide site binding tankyrase inhibitors.
Eur.J.Med.Chem., 288, 2025
|
|
8S2F
 
 | |
8RMX
 
 | | Transglutaminase 3 in complex with DH patient-derived Fab DH63-A02 | | Descriptor: | Antibody Fab fragment heavy chain IGHV2-5, Antibody Fab fragment light chain IGKV4-1, CALCIUM ION, ... | | Authors: | Heggelund, J.E, Sollid, L.M. | | Deposit date: | 2024-01-09 | | Release date: | 2025-03-12 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Enzyme-activating B-cell receptors boost antigen presentation to pathogenic T cells in gluten-sensitive autoimmunity.
Nat Commun, 16, 2025
|
|
