1C6T
 
 | |
1C6V
 
 | | SIV INTEGRASE (CATALYTIC DOMAIN + DNA BIDING DOMAIN COMPRISING RESIDUES 50-293) MUTANT WITH PHE 185 REPLACED BY HIS (F185H) | | Descriptor: | PROTEIN (SIU89134), PROTEIN (SIV INTEGRASE) | | Authors: | Chen, Z, Yan, Y, Munshi, S, Li, Y, Zruygay-Murphy, J, Xu, B, Witmer, M, Felock, P, Wolfe, A, Sardana, V, Emini, E.A, Hazuda, D, Kuo, L.C. | | Deposit date: | 1999-12-21 | | Release date: | 2000-12-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray structure of simian immunodeficiency virus integrase containing the core and C-terminal domain (residues 50-293)--an initial glance of the viral DNA binding platform.
J.Mol.Biol., 296, 2000
|
|
1C6W
 
 | | MAUROCALCIN FROM SCORPIO MAURUS | | Descriptor: | MAUROCALCIN | | Authors: | Mosbah, A, Kharrat, R, Fajloun, Z, Renisio, J.-G, Blanc, E, Sabatier, J.-M, Ayeb, M, Darbon, H. | | Deposit date: | 1999-12-23 | | Release date: | 2000-09-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A new fold in the scorpion toxin family, associated with an activity on a ryanodine-sensitive calcium channel.
Proteins, 40, 2000
|
|
1C6X
 
 | |
1C6Y
 
 | |
1C6Z
 
 | | ALTERNATE BINDING SITE FOR THE P1-P3 GROUP OF A CLASS OF POTENT HIV-1 PROTEASE INHIBITORS AS A RESULT OF CONCERTED STRUCTURAL CHANGE IN 80'S LOOP. | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, PROTEIN (PROTEASE) | | Authors: | Munshi, S. | | Deposit date: | 1999-12-28 | | Release date: | 2000-12-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An alternate binding site for the P1-P3 group of a class of potent HIV-1 protease inhibitors as a result of concerted structural change in the 80s loop of the protease.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1C70
 
 | |
1C72
 
 | | TYR115, GLN165 AND TRP209 CONTRIBUTE TO THE 1,2-EPOXY-3-(P-NITROPHENOXY)PROPANE CONJUGATING ACTIVITIES OF GLUTATHIONE S-TRANSFERASE CGSTM1-1 | | Descriptor: | 1-HYDROXY-2-S-GLUTATHIONYL-3-PARA-NITROPHENOXY-PROPANE, PROTEIN (GLUTATHIONE S-TRANSFERASE) | | Authors: | Chern, M.K, Wu, T.C, Hsieh, C.H, Chou, C.C, Liu, L.F, Kuan, I.C, Yeh, Y.H, Hsiao, C.D, Tam, M.F. | | Deposit date: | 2000-02-02 | | Release date: | 2000-08-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Tyr115, gln165 and trp209 contribute to the 1, 2-epoxy-3-(p-nitrophenoxy)propane-conjugating activity of glutathione S-transferase cGSTM1-1.
J.Mol.Biol., 300, 2000
|
|
1C74
 
 | | Structure of the double mutant (K53,56M) of phospholipase A2 | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Sekar, K, Tsai, M.D, Jain, M.K, Ramakumar, S. | | Deposit date: | 2000-01-22 | | Release date: | 2000-07-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of the anionic interface preference and k*cat activation of pancreatic phospholipase A2.
Biochemistry, 39, 2000
|
|
1C75
 
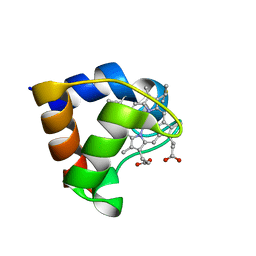 | | 0.97 A "AB INITIO" CRYSTAL STRUCTURE OF CYTOCHROME C-553 FROM BACILLUS PASTEURII | | Descriptor: | CYTOCHROME C-553, HEME C | | Authors: | Benini, S, Ciurli, S, Rypniewski, W.R, Wilson, K.S. | | Deposit date: | 2000-02-09 | | Release date: | 2000-03-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Crystal structure of oxidized Bacillus pasteurii cytochrome c553 at 0.97-A resolution.
Biochemistry, 39, 2000
|
|
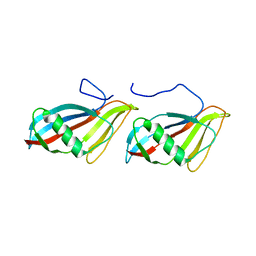
1C76
 
 | | STAPHYLOKINASE (SAK) MONOMER | | Descriptor: | STAPHYLOKINASE | | Authors: | Rao, Z, Jiang, F, Liu, Y, Zhang, X, Chen, Y, Bartlam, M, Song, H, Ding, Y. | | Deposit date: | 2000-02-01 | | Release date: | 2000-08-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Staphylokinase Dimer Offers New Clue to Reduction of Immunogenicity
To be published
|
|
1C77
 
 | | STAPHYLOKINASE (SAK) DIMER | | Descriptor: | STAPHYLOKINASE | | Authors: | Rao, Z, Jiang, F, Liu, Y, Zhang, X, Chen, Y, Bartlam, M, Song, H, Ding, Y. | | Deposit date: | 2000-02-01 | | Release date: | 2000-08-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a staphylokinase: variant a model for reduced antigenicity.
Eur.J.Biochem., 269, 2002
|
|
1C78
 
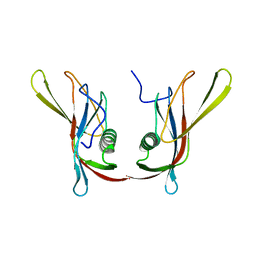 | | STAPHYLOKINASE (SAK) DIMER | | Descriptor: | STAPHYLOKINASE | | Authors: | Rao, Z, Jiang, F, Liu, Y, Zhang, X, Chen, Y, Bartlam, M, Song, H, Ding, Y. | | Deposit date: | 2000-02-01 | | Release date: | 2000-08-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a staphylokinase: variant a model for reduced antigenicity.
Eur.J.Biochem., 269, 2002
|
|
1C79
 
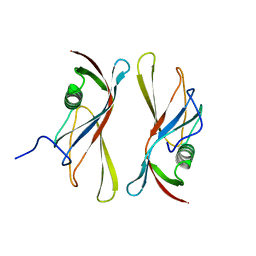 | | STAPHYLOKINASE (SAK) DIMER | | Descriptor: | STAPHYLOKINASE | | Authors: | Rao, Z, Jiang, F, Liu, Y, Zhang, X, Chen, Y, Bartlam, M, Song, H, Ding, Y. | | Deposit date: | 2000-02-01 | | Release date: | 2000-08-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a staphylokinase: variant a model for reduced antigenicity.
Eur.J.Biochem., 269, 2002
|
|
1C7B
 
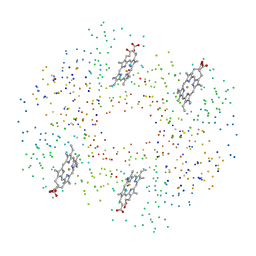 | | DEOXY RHB1.0 (RECOMBINANT HEMOGLOBIN) | | Descriptor: | PROTEIN (DEOXYHEMOGLOBIN (ALPHA CHAIN)), PROTEIN (DEOXYHEMOGLOBIN (BETA CHAIN)), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Brucker, E.A. | | Deposit date: | 2000-02-09 | | Release date: | 2000-06-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Genetically crosslinked hemoglobin: a structural study.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1C7C
 
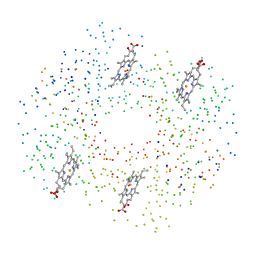 | | DEOXY RHB1.1 (RECOMBINANT HEMOGLOBIN) | | Descriptor: | PROTEIN (DEOXYHEMOGLOBIN (ALPHA CHAIN)), PROTEIN (DEOXYHEMOGLOBIN (BETA CHAIN)), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Brucker, E.A. | | Deposit date: | 2000-02-09 | | Release date: | 2000-06-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Genetically crosslinked hemoglobin: a structural study.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1C7D
 
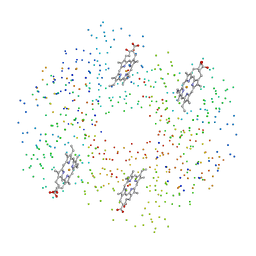 | | DEOXY RHB1.2 (RECOMBINANT HEMOGLOBIN) | | Descriptor: | PROTEIN (DEOXYHEMOGLOBIN (ALPHA CHAIN)), PROTEIN (DEOXYHEMOGLOBIN (BETA CHAIN)), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Brucker, E.A. | | Deposit date: | 2000-02-09 | | Release date: | 2000-06-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Genetically crosslinked hemoglobin: a structural study.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1C7E
 
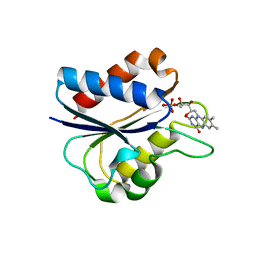 | | D95E HYDROQUINONE FLAVODOXIN MUTANT FROM D. VULGARIS | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | McCarthy, A, Walsh, M, Higgins, T, D'Arcy, D. | | Deposit date: | 2000-02-16 | | Release date: | 2000-08-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystallographic Investigation of the Role of Aspartate 95 in the Modulation of the Redox Potentials Of Desulfovibrio Vulgaris Flavodoxin
Biochemistry, 41, 2002
|
|
1C7F
 
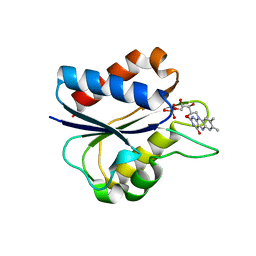 | | D95E OXIDIZED FLAVODOXIN MUTANT FROM D. VULGARIS | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | McCarthy, A, Walsh, M, Higgins, T, D'Arcy, D. | | Deposit date: | 2000-02-11 | | Release date: | 2000-08-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic Investigation of the Role of Aspartate 95 in the Modulation of the Redox Potentials Of Desulfovibrio Vulgaris Flavodoxin
Biochemistry, 41, 2002
|
|
1C7G
 
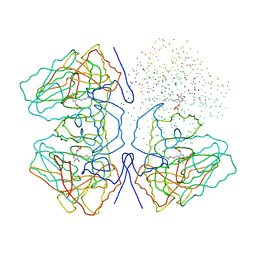 | |
1C7H
 
 | | CRYSTAL STRUCTURE OF A MUTANT R75A IN KETOSTEROID ISOMERASE FROM PSEDOMONAS PUTIDA BIOTYPE B | | Descriptor: | DELTA-5-3-KETOSTEROID ISOMERASE | | Authors: | Nam, G.H, Kim, D.H, Jang, D.S, Choi, G, Ha, N.C, Oh, B.H, Choi, K.Y. | | Deposit date: | 2000-02-19 | | Release date: | 2000-04-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Roles of active site aromatic residues in catalysis by ketosteroid isomerase from Pseudomonas putida biotype B.
Biochemistry, 38, 1999
|
|
1C7I
 
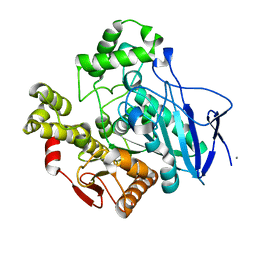 | | THERMOPHYLIC PNB ESTERASE | | Descriptor: | CALCIUM ION, PROTEIN (PARA-NITROBENZYL ESTERASE) | | Authors: | Spiller, B, Gershenson, A, Arnold, F, Stevens, R. | | Deposit date: | 2000-02-21 | | Release date: | 2000-03-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural view of evolutionary divergence.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1C7J
 
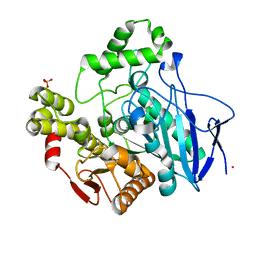 | | PNB ESTERASE 56C8 | | Descriptor: | POTASSIUM ION, PROTEIN (PARA-NITROBENZYL ESTERASE), SULFATE ION | | Authors: | Spiller, B, Gershenson, A, Arnold, F, Stevens, R.C. | | Deposit date: | 2000-02-21 | | Release date: | 2000-03-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A structural view of evolutionary divergence.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1C7K
 
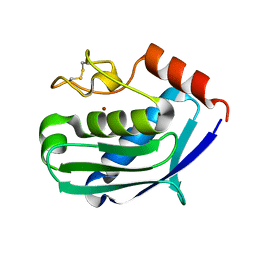 | | CRYSTAL STRUCTURE OF THE ZINC PROTEASE | | Descriptor: | CALCIUM ION, ZINC ENDOPROTEASE, ZINC ION | | Authors: | Kurisu, G, Harada, S, Kai, Y. | | Deposit date: | 2000-02-19 | | Release date: | 2001-04-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure of the zinc-binding site in the crystal structure of a zinc endoprotease from Streptomyces caespitosus at 1 A resolution.
J.Inorg.Biochem., 82, 2000
|
|
1C7M
 
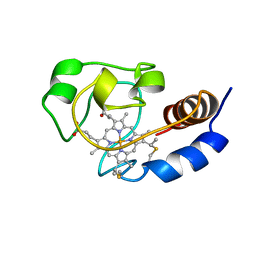 | | SOLUTION STRUCTURE OF THE FUNCTIONAL DOMAIN OF PARACOCCUS DENITRIFICANS CYTOCHROME C552 IN THE REDUCED STATE | | Descriptor: | HEME C, PROTEIN (CYTOCHROME C552) | | Authors: | Pristovsek, P, Luecke, C, Reincke, B, Ludwig, B, Rueterjans, H. | | Deposit date: | 2000-03-03 | | Release date: | 2000-07-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the functional domain of Paracoccus denitrificans cytochrome c552 in the reduced state.
Eur.J.Biochem., 267, 2000
|
|
