6T88
 
 | |
6T85
 
 | | Urocanate reductase in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Venskutonyte, R, Lindkvist-Petersson, K. | | Deposit date: | 2019-10-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural characterization of the microbial enzyme urocanate reductase mediating imidazole propionate production.
Nat Commun, 12, 2021
|
|
5O08
 
 | | Crystal structure of Phosphopantetheine adenylyltransferase from Mycobacterium abcessus in complex with Dephospho-coenzyme A | | Descriptor: | 1,2-ETHANEDIOL, DEPHOSPHO COENZYME A, Phosphopantetheine adenylyltransferase | | Authors: | Thomas, S.E, Kim, S.Y, Mendes, V, Blaszczyk, M, Blundell, T.L. | | Deposit date: | 2017-05-16 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | Structural Biology and the Design of New Therapeutics: From HIV and Cancer to Mycobacterial Infections: A Paper Dedicated to John Kendrew.
J. Mol. Biol., 429, 2017
|
|
5O0D
 
 | | Crystal structure of Phosphopantetheine adenylyltransferase from Mycobacterium abcessus in complex with 3-Phenoxymandelic acid (Fragment 4) | | Descriptor: | (2~{S})-2-oxidanyl-2-(3-phenoxyphenyl)ethanoic acid, Phosphopantetheine adenylyltransferase | | Authors: | Thomas, S.E, Kim, S.Y, Mendes, V, Blaszczyk, M, Blundell, T.L. | | Deposit date: | 2017-05-16 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.539 Å) | | Cite: | Structural Biology and the Design of New Therapeutics: From HIV and Cancer to Mycobacterial Infections: A Paper Dedicated to John Kendrew.
J. Mol. Biol., 429, 2017
|
|
7ARA
 
 | | Rhinovirus A2 2A protease in complex with zVAM.fmk | | Descriptor: | (phenylmethyl) ~{N}-[(2~{R})-3-methyl-1-[[(2~{S})-1-[[(3~{S})-1-methylsulfanyl-4-oxidanylidene-pentan-3-yl]amino]-1-oxidanylidene-propan-2-yl]amino]-1-oxidanylidene-butan-2-yl]carbamate, (phenylmethyl) ~{N}-[(2~{S})-3-methyl-1-[[(2~{R})-1-[[(3~{R})-1-methylsulfanyl-4-oxidanylidene-pentan-3-yl]amino]-1-oxidanylidene-propan-2-yl]amino]-1-oxidanylidene-butan-2-yl]carbamate, 2A protease, ... | | Authors: | Deutschmann-Olek, K.M, Skern, T, Bezerra, G.A, Yue, W.W. | | Deposit date: | 2020-10-23 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.243 Å) | | Cite: | Defining substrate selection by rhinoviral 2A proteinase through its crystal structure with the inhibitor zVAM.fmk.
Virology, 562, 2021
|
|
6T1V
 
 | | Structure of PPARg H494Y mutant in complex with GW1929 | | Descriptor: | (2~{S})-3-[4-[2-[methyl(pyridin-2-yl)amino]ethoxy]phenyl]-2-[[2-(phenylcarbonyl)phenyl]amino]propanoic acid, Peroxisome proliferator-activated receptor gamma, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha | | Authors: | Rochel, N. | | Deposit date: | 2019-10-06 | | Release date: | 2021-04-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure of a PPARg mutant complex
To Be Published
|
|
6T1S
 
 | | PPAR mutant | | Descriptor: | (2~{S})-3-[4-[2-[methyl(pyridin-2-yl)amino]ethoxy]phenyl]-2-[[2-(phenylcarbonyl)phenyl]amino]propanoic acid, Peroxisome proliferator-activated receptor gamma, SULFATE ION | | Authors: | Rochel, N. | | Deposit date: | 2019-10-05 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of PPARg mutant
To Be Published
|
|
6T2A
 
 | |
5O0S
 
 | | Crystal structure of txGH116 (beta-glucosidase from Thermoanaerobacterium xylolyticum) in complex with unreacted beta Cyclophellitol Cyclosulfate probe ME711 | | Descriptor: | (3~{a}~{S},4~{R},5~{S},6~{R},7~{R},7~{a}~{R})-7-(hydroxymethyl)-2,2-bis(oxidanylidene)-3~{a},4,5,6,7,7~{a}-hexahydrobenzo[d][1,3,2]dioxathiole-4,5,6-triol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Wu, L, Offen, W.A, Breen, I.Z, Davies, G.J. | | Deposit date: | 2017-05-16 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | 1,6-Cyclophellitol Cyclosulfates: A New Class of Irreversible Glycosidase Inhibitor.
ACS Cent Sci, 3, 2017
|
|
6TE3
 
 | | Crystal Structure of full-length Human Lysyl Hydroxylase LH3 - Cocrystal with Fe2+, Mn2+, UDP | | Descriptor: | 2-OXOGLUTARIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chiapparino, A, De Giorgi, F, Scietti, L, Faravelli, S, Roscioli, T, Forneris, F. | | Deposit date: | 2019-11-11 | | Release date: | 2021-05-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of Regulatory Molecular 'Hot Spots' for LH/PLOD Collagen Glycosyltransferase Activity
Int J Mol Sci, 2023
|
|
5NX2
 
 | | Crystal structure of thermostabilised full-length GLP-1R in complex with a truncated peptide agonist at 3.7 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glucagon-like peptide 1 receptor, ... | | Authors: | Rappas, M, Jazayeri, A, Brown, A.J.H, Kean, J, Errey, J.C, Robertson, N, Fiez-Vandal, C, Andrews, S.P, Congreve, M, Bortolato, A, Mason, J.S, Baig, A.H, Teobald, I, Dore, A.S, Weir, M, Cooke, R.M, Marshall, F.H. | | Deposit date: | 2017-05-09 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of the GLP-1 receptor bound to a peptide agonist.
Nature, 546, 2017
|
|
6TEC
 
 | | Crystal Structure of full-length Human Lysyl Hydroxylase LH3 - Cocrystal with Fe2+, Mn2+, UDP-Xylose | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chiapparino, A, De Giorgi, F, Scietti, L, Faravelli, S, Roscioli, T, Forneris, F. | | Deposit date: | 2019-11-11 | | Release date: | 2021-05-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of Regulatory Molecular 'Hot Spots' for LH/PLOD Collagen Glycosyltransferase Activity
Int J Mol Sci, 2023
|
|
7B0I
 
 | | Structure of a minimal SF3B core in complex with spliceostatin A (form II) | | Descriptor: | PHD finger-like domain-containing protein 5A, Splicing factor 3B subunit 1, Splicing factor 3B subunit 3,Splicing factor 3B subunit 3,Splicing factor 3B subunit 3, ... | | Authors: | Cretu, C, Pena, V. | | Deposit date: | 2020-11-19 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of intron selection by U2 snRNP in the presence of covalent inhibitors.
Nat Commun, 12, 2021
|
|
6TES
 
 | | Crystal Structure of full-length Human Lysyl Hydroxylase LH3 - Cocrystal with Fe2+, Mn2+, UDP-Glucuronic Acid | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chiapparino, A, De Giorgi, F, Scietti, L, Faravelli, S, Roscioli, T, Forneris, F. | | Deposit date: | 2019-11-12 | | Release date: | 2021-05-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of Regulatory Molecular 'Hot Spots' for LH/PLOD Collagen Glycosyltransferase Activity
Int J Mol Sci, 2023
|
|
5OBZ
 
 | | low resolution structure of the p34ct/p44ct minimal complex | | Descriptor: | Putative transcription factor, ZINC ION | | Authors: | Schoenwetter, E, Koelmel, W, Schmitt, D.R, Kuper, J, Kisker, C. | | Deposit date: | 2017-06-29 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | The intricate network between the p34 and p44 subunits is central to the activity of the transcription/DNA repair factor TFIIH.
Nucleic Acids Res., 45, 2017
|
|
7AH8
 
 | | NF-Y bound to suramin inhibitor | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFON IC ACID, CITRATE ANION, GLYCEROL, ... | | Authors: | Nardone, V, Chaves-Sanjuan, A, Lapi, M, Nardini, M. | | Deposit date: | 2020-09-24 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.70001364 Å) | | Cite: | Structural Basis of Inhibition of the Pioneer Transcription Factor NF-Y by Suramin.
Cells, 9, 2020
|
|
6TEU
 
 | | Crystal Structure of full-length Human Lysyl Hydroxylase LH3 - Val80Lys mutant - Cocrystal with Fe2+, Mn2+ | | Descriptor: | 2-OXOGLUTARIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chiapparino, A, De Giorgi, F, Scietti, L, Faravelli, S, Roscioli, T, Forneris, F. | | Deposit date: | 2019-11-12 | | Release date: | 2021-05-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Identification of Regulatory Molecular 'Hot Spots' for LH/PLOD Collagen Glycosyltransferase Activity
Int J Mol Sci, 2023
|
|
7AT5
 
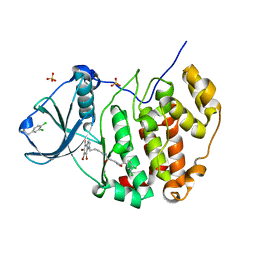 | | Structure of protein kinase ck2 catalytic subunit (csnk2a1 gene product) in complex with the bivalent inhibitor KN2 | | Descriptor: | 1,2-ETHANEDIOL, 2-(3,4-dichlorophenyl)ethanamine, Casein kinase II subunit alpha, ... | | Authors: | Lindenblatt, D, Applegate, V, Nickelsen, A, Klussmann, M, Neundorf, I, Goetz, C, Jose, J, Niefind, K. | | Deposit date: | 2020-10-29 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Molecular Plasticity of Crystalline CK2 alpha ' Leads to KN2, a Bivalent Inhibitor of Protein Kinase CK2 with Extraordinary Selectivity.
J.Med.Chem., 65, 2022
|
|
6TEZ
 
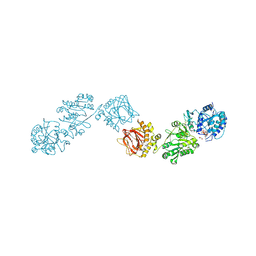 | | Crystal Structure of full-length Human Lysyl Hydroxylase LH3 - Val80Lys mutant - Cocrystal with Fe2+, Mn2+, UDP-Glucuronic Acid | | Descriptor: | 2-OXOGLUTARIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chiapparino, A, De Giorgi, F, Scietti, L, Faravelli, S, Roscioli, T, Forneris, F. | | Deposit date: | 2019-11-12 | | Release date: | 2021-05-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of Regulatory Molecular 'Hot Spots' for LH/PLOD Collagen Glycosyltransferase Activity
Int J Mol Sci, 2023
|
|
7ALT
 
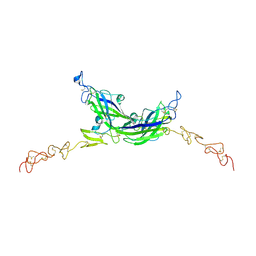 | | Structure of Drosophila Serrate C2-DSL-EGF1-EGF2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Suckling, R, Johnson, S, Lea, S.M. | | Deposit date: | 2020-10-07 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The conserved C2 phospholipid-binding domain in Delta contributes to robust Notch signalling.
Embo Rep., 22, 2021
|
|
6TEX
 
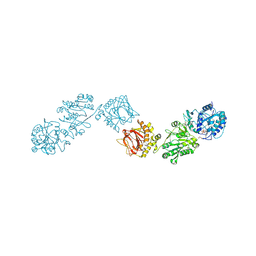 | | Crystal Structure of full-length Human Lysyl Hydroxylase LH3 - Val80Lys mutant - Cocrystal with Fe2+, Mn2+, UDP-Glucose | | Descriptor: | 2-OXOGLUTARIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chiapparino, A, De Giorgi, F, Scietti, L, Faravelli, S, Roscioli, T, Forneris, F. | | Deposit date: | 2019-11-12 | | Release date: | 2021-05-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of Regulatory Molecular 'Hot Spots' for LH/PLOD Collagen Glycosyltransferase Activity
Int J Mol Sci, 2023
|
|
5O2U
 
 | | Llama VHH in complex with p24 | | Descriptor: | Capsid protein p24, VHH 59H10 | | Authors: | Caillat, C, Verrips, T, Weissenhorn, W. | | Deposit date: | 2017-05-22 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Unravelling the Molecular Basis of High Affinity Nanobodies against HIV p24: In Vitro Functional, Structural, and in Silico Insights.
ACS Infect Dis, 3, 2017
|
|
5OCC
 
 | |
5O35
 
 | | Structure of complement proteins complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C3, Complement factor H,Complement factor H | | Authors: | Xue, X, Wu, J, Forneris, F, Gros, P. | | Deposit date: | 2017-05-23 | | Release date: | 2017-06-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Regulator-dependent mechanisms of C3b processing by factor I allow differentiation of immune responses.
Nat. Struct. Mol. Biol., 24, 2017
|
|
6TKB
 
 | | ChiLob 7/4 H2 HC-C224S F(ab')2 | | Descriptor: | Chilob 7/4 H2 heavy chain C224S, Chilob 7/4 H2 kappa chain | | Authors: | Orr, C.M, Fisher, H, Tews, I. | | Deposit date: | 2019-11-28 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hinge disulfides in human IgG2 CD40 antibodies modulate receptor signaling by regulation of conformation and flexibility.
Sci Immunol, 7, 2022
|
|
