4LCJ
 
 | | CtBP2 in complex with substrate MTOB | | Descriptor: | 4-(METHYLSULFANYL)-2-OXOBUTANOIC ACID, C-terminal-binding protein 2, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Hilbert, B.J, Schiffer, C.A, Royer Jr, W.E. | | Deposit date: | 2013-06-21 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Crystal structures of human CtBP in complex with substrate MTOB reveal active site features useful for inhibitor design.
Febs Lett., 588, 2014
|
|
1KRP
 
 | | DNA polymerase I Klenow fragment (E.C.2.7.7.7) mutant/DNA complex | | Descriptor: | DNA (5'-D(P*TP*TP*PST)-3'), PROTEIN (DNA POLYMERASE I KLENOW FRAGMENT (E.C.2.7.7.7)), ZINC ION | | Authors: | Brautigam, C.A, Steitz, T.A. | | Deposit date: | 1997-08-19 | | Release date: | 1998-02-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural principles for the inhibition of the 3'-5' exonuclease activity of Escherichia coli DNA polymerase I by phosphorothioates.
J.Mol.Biol., 277, 1998
|
|
1KSP
 
 | | DNA polymerase I Klenow fragment (E.C.2.7.7.7) mutant/DNA complex | | Descriptor: | DNA (5'-D(P*TP*TP*PST)-3'), PROTEIN (DNA POLYMERASE I-KLENOW FRAGMENT (E.C.2.7.7.7)), ZINC ION | | Authors: | Brautigam, C.A, Steitz, T.A. | | Deposit date: | 1997-08-19 | | Release date: | 1998-02-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural principles for the inhibition of the 3'-5' exonuclease activity of Escherichia coli DNA polymerase I by phosphorothioates.
J.Mol.Biol., 277, 1998
|
|
3CTN
 
 | | STRUCTURE OF CALCIUM-SATURATED CARDIAC TROPONIN C, NMR, 30 STRUCTURES | | Descriptor: | CALCIUM ION, TROPONIN C | | Authors: | Sia, S.K, Li, M.X, Spyracopoulos, L, Gagne, S.M, Liu, W, Putkey, J.A, Sykes, B.D. | | Deposit date: | 1997-05-08 | | Release date: | 1998-05-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of cardiac muscle troponin C unexpectedly reveals a closed regulatory domain.
J.Biol.Chem., 272, 1997
|
|
1LM0
 
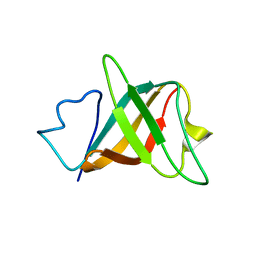 | | Solution structure and characterization of the heme chaperone CcmE | | Descriptor: | cytochrome c maturation protein E | | Authors: | Arnesano, F, Banci, L, Barker, P.D, Bertini, I, Rosato, A, Su, X.C, Viezzoli, M.S. | | Deposit date: | 2002-04-30 | | Release date: | 2002-12-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and characterization of the heme chaperone CcmE
Biochemistry, 41, 2002
|
|
9MX8
 
 | |
4LOH
 
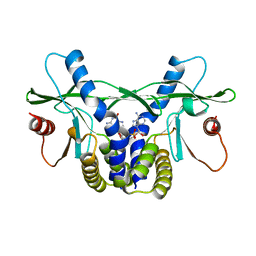 | | Crystal structure of hSTING(H232) in complex with c[G(2',5')pA(3',5')p] | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Gao, P, Patel, D.J. | | Deposit date: | 2013-07-12 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Function Analysis of STING Activation by c[G(2',5')pA(3',5')p] and Targeting by Antiviral DMXAA.
Cell(Cambridge,Mass.), 154, 2013
|
|
4LOJ
 
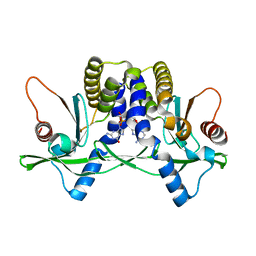 | | Crystal structure of mSting in complex with c[G(2',5')pA(3',5')p] | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Gao, P, Patel, D.J. | | Deposit date: | 2013-07-12 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure-Function Analysis of STING Activation by c[G(2',5')pA(3',5')p] and Targeting by Antiviral DMXAA.
Cell(Cambridge,Mass.), 154, 2013
|
|
3SBG
 
 | |
5K65
 
 | | Crystal structure of VEGF binding IgG1-Fc (Fcab CT6) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Humm, A, Lobner, E, Kitzmuller, M, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2016-05-24 | | Release date: | 2017-09-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two-faced Fcab prevents polymerization with VEGF and reveals thermodynamics and the 2.15 angstrom crystal structure of the complex.
MAbs, 9, 2017
|
|
3S7W
 
 | | Structure of the TvNiRb form of Thioalkalivibrio nitratireducens cytochrome c nitrite reductase with an oxidized Gln360 in a complex with hydroxylamine | | Descriptor: | AZIDE ION, CALCIUM ION, Eight-heme nitrite reductase, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2011-05-27 | | Release date: | 2011-07-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure of the TvNiRb form of Thioalkalivibrio nitratireducens cytochrome c nitrite reductase with an oxidized Gln360 in a complex with hydroxylamine
To be Published
|
|
8HKD
 
 | |
2V08
 
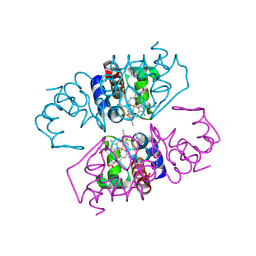 | | Structure of wild-type Phormidium laminosum cytochrome c6 | | Descriptor: | CHLORIDE ION, CYTOCHROME C6, HEME C, ... | | Authors: | Worrall, J.A.R, Schlarb-Ridley, B.G, Reda, T, Marcaida, M.J, Moorlen, R.J, Wastl, J, Hirst, J, Bendall, D.S, Luisi, B.F, Howe, C.J. | | Deposit date: | 2007-05-10 | | Release date: | 2007-07-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Modulation of heme redox potential in the cytochrome c6 family.
J. Am. Chem. Soc., 129, 2007
|
|
5DCK
 
 | |
5XGN
 
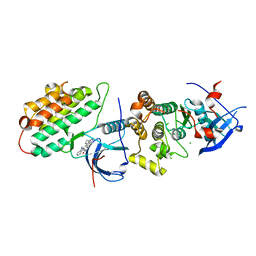 | | Crystal structure of EGFR 696-1022 T790M/C797S in complex with Go6976 | | Descriptor: | 12-(2-Cyanoethyl)-6,7,12,13-tetrahydro-13-methyl-5-oxo-5H-indolo[2,3-a]pyrrolo[3,4-c]carbazole, CHLORIDE ION, Epidermal growth factor receptor | | Authors: | Kong, L.L, Yun, C.H. | | Deposit date: | 2017-04-14 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural pharmacological studies on EGFR T790M/C797S.
Biochem. Biophys. Res. Commun., 488, 2017
|
|
5XR8
 
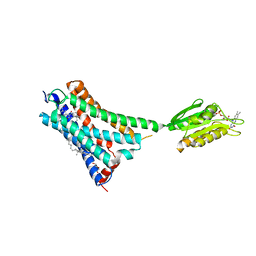 | | Crystal structure of the human CB1 in complex with agonist AM841 | | Descriptor: | (6~{a}~{R},9~{R},10~{a}~{R})-9-(hydroxymethyl)-3-(8-isothiocyanato-2-methyl-octan-2-yl)-6,6-dimethyl-6~{a},7,8,9,10,10~{a}-hexahydrobenzo[c]chromen-1-ol, CHOLESTEROL, Cannabinoid receptor 1,Flavodoxin,Cannabinoid receptor 1, ... | | Authors: | Hua, T, Vemuri, K, Nikas, P.S, Laprairie, R.B, Wu, Y, Qu, L, Pu, M, Korde, A, Shan, J, Ho, J.H, Han, G.W, Ding, K, Li, X, Liu, H, Hanson, M.A, Zhao, S, Bohn, L.M, Makriyannis, A, Stevens, R.C, Liu, Z.J. | | Deposit date: | 2017-06-07 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structures of agonist-bound human cannabinoid receptor CB1.
Nature, 547, 2017
|
|
5D8K
 
 | |
2V07
 
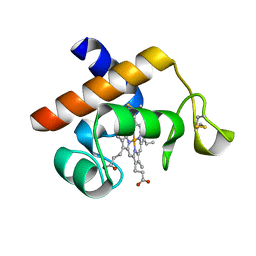 | | Structure of the Arabidopsis thaliana cytochrome c6A V52Q variant | | Descriptor: | CYTOCHROME C6, HEME C | | Authors: | Worrall, J.A.R, Schlarb-Ridley, B.G, Reda, T, Marcaida, M.J, Moorlen, R.J, Wastl, J, Hirst, J, Bendall, D.S, Luisi, B.F, Howe, C.J. | | Deposit date: | 2007-05-09 | | Release date: | 2007-07-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Modulation of heme redox potential in the cytochrome c6 family.
J. Am. Chem. Soc., 129, 2007
|
|
5DDV
 
 | |
4URS
 
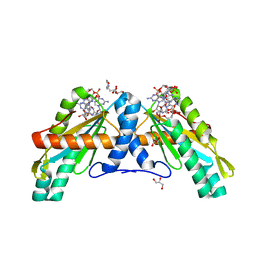 | | Crystal Structure of GGDEF domain from T.maritima | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), DIGUANYLATE CYCLASE, ... | | Authors: | Deepthi, A, Liew, C.W, Liang, Z.X, Swaminathan, K, Lescar, J. | | Deposit date: | 2014-07-02 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure of a Diguanylate Cyclase from Thermotoga Maritima: Insights Into Activation, Feedback Inhibition and Thermostability
Plos One, 9, 2014
|
|
3CU4
 
 | |
9MT0
 
 | |
4UZC
 
 | |
3SLE
 
 | |
1KNZ
 
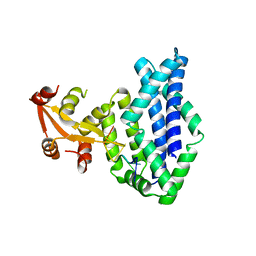 | | Recognition of the rotavirus mRNA 3' consensus by an asymmetric NSP3 homodimer | | Descriptor: | 5'-R(*UP*GP*AP*CP*C)-3', Nonstructural RNA-binding Protein 34 | | Authors: | Deo, R.C, Groft, C.M, Rajashankar, K.R, Burley, S.K. | | Deposit date: | 2001-12-19 | | Release date: | 2002-01-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Recognition of the rotavirus mRNA 3' consensus by an asymmetric NSP3 homodimer.
Cell(Cambridge,Mass.), 108, 2002
|
|
