6OH8
 
 | |
6P1K
 
 | | Cryo-EM structure of Escherichia coli sigma70 bound RNAP polymerase holoenzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | Deposit date: | 2019-05-20 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | E. coliTraR allosterically regulates transcription initiation by altering RNA polymerase conformation.
Elife, 8, 2019
|
|
6F86
 
 | | Crystal Structure of E. coli GyraseB 24kDa in complex with 4-(4-bromo-1H-pyrazol-1-yl)-6-[(ethylcarbamoyl)amino]-N-(pyridin-3-yl)pyridine-3-carboxamide | | Descriptor: | 4-(4-bromanylpyrazol-1-yl)-6-(ethylcarbamoylamino)-~{N}-pyridin-3-yl-pyridine-3-carboxamide, DNA gyrase subunit B | | Authors: | Narramore, S.K, Stevenson, C.E.M, Lawson, D.M, Maxwell, A, Fishwick, C.W.G. | | Deposit date: | 2017-12-12 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New insights into the binding mode of pyridine-3-carboxamide inhibitors of E. coli DNA gyrase.
Bioorg.Med.Chem., 27, 2019
|
|
6F94
 
 | | Crystal Structure of E. coli GyraseB 24kDa in complex with 6-[(ethylcarbamoyl)amino]-4-[(3-methyphenyl)amino]-N-(3-methyphenyl)pyridine-3-carboxamide | | Descriptor: | 6-(ethylcarbamoylamino)-~{N}-(3-methylphenyl)-4-[(3-methylphenyl)amino]pyridine-3-carboxamide, DNA gyrase subunit B | | Authors: | Narramore, S.K, Stevenson, C.E.M, Lawson, D.M, Maxwell, A, Fishwick, C.W.G. | | Deposit date: | 2017-12-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | New insights into the binding mode of pyridine-3-carboxamide inhibitors of E. coli DNA gyrase.
Bioorg.Med.Chem., 27, 2019
|
|
2KHO
 
 | |
2KU0
 
 | | Inhibitor Induced Structural Change in the HCV IRES Domain IIa RNA | | Descriptor: | (7S)-7-[(dimethylamino)methyl]-1-[3-(dimethylamino)propyl]-7,8-dihydro-1H-furo[3,2-e]benzimidazol-2-amine, HCV IRES Domain IIa RNA | | Authors: | Paulsen, R.B, Seth, P.P, Swayze, E.E, Griffey, R.H, Skalicky, J.J, Cheatham III, T.E, Davis, D.R. | | Deposit date: | 2010-02-10 | | Release date: | 2010-04-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Inhibitor-induced structural change in the HCV IRES domain IIa RNA.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6PNR
 
 | | A GH31 family sulfoquinovosidase from E. rectale in complex with aza-sugar inhibitor IFGSQ | | Descriptor: | Alpha-glucosidase, SULFATE ION, [(3~{S},4~{R},5~{R})-4,5-bis(oxidanyl)piperidin-3-yl]methanesulfonic acid | | Authors: | Jarva, M.A, Lingford, J.P, John, A, Goddard-Borger, E.D. | | Deposit date: | 2019-07-03 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A GH31 family sulfoquinovosidase from E. rectale in complex with aza-sugar inhibitor IFGSQ
To Be Published
|
|
2L61
 
 | | Protein and metal cluster structure of the wheat metallothionein domain g-Ec-1. The second part of the puzzle. | | Descriptor: | CADMIUM ION, EC protein I/II | | Authors: | Loebus, J, Peroza, E.A, Bluethgen, N, Fox, T, Meyer-Klaucke, W, Zerbe, O, Freisinger, E. | | Deposit date: | 2010-11-12 | | Release date: | 2011-05-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Protein and metal cluster structure of the wheat metallothionein domain gamma-E(c)-1: the second part of the puzzle.
J.Biol.Inorg.Chem., 16, 2011
|
|
4XYC
 
 | | NANOMOLAR INHIBITORS OF MYCOBACTERIUM TUBERCULOSIS GLUTAMINE SYNTHETASE 1: SYNTHESIS, BIOLOGICAL EVALUATION AND X-RAY CRYSTALLOGRAPHIC STUDIES | | Descriptor: | 9-phenyl-4H-imidazo[1,2-a]indeno[1,2-e]pyrazin-4-one, Glutamine synthetase 1 | | Authors: | Couturier, C, Silve, S, Morales, R, Ppessegue, B, Llopart, S, Nair, A, Bauer, A, Scheiper, B, poeverlein, c, Ganzhorn, A, Lagrange, S, Bacque, E. | | Deposit date: | 2015-02-02 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Nanomolar inhibitors of Mycobacterium tuberculosis glutamine synthetase 1: Synthesis, biological evaluation and X-ray crystallographic studies.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
6XE3
 
 | | Salmonella typhimurium Tryptophan Synthase beta-S377A mutant in complex with inhibitor F9 at the enzyme alpha-site, cesium ion at the metal coordination site and carbanion III E(C3) at the enzyme beta-site. | | Descriptor: | (2E)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]-3-[(2-hydroxyphenyl)amino]propanoic acid, 1,2-ETHANEDIOL, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2020-06-11 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Salmonella typhimurium Tryptophan Synthase beta-S377A mutant in complex with inhibitor F9 at the enzyme alpha-site, cesium ion at the metal coordination site and carbanion III E(C3) at the enzyme beta-site.
To be Published
|
|
4YF1
 
 | | 1.85 angstrom crystal structure of lmo0812 from Listeria monocytogenes EGD-e | | Descriptor: | CITRATE ANION, Lmo0812 protein, SODIUM ION | | Authors: | Krishna, S.N, Light, S.H, Filippova, E.V, Minasov, G, Kiryukhina, O, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-02-24 | | Release date: | 2015-03-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 angstrom crystal structure of lmo0812 from Listeria monocytogenes EGD-e
To Be Published
|
|
8R0H
 
 | | Pim1 in complex with (E)-3-(2-(thiophen-2-yl)vinyl)-3,4-dihydroquinoxalin-2(1H)-one and Pimtide | | Descriptor: | 3-(2-thiophen-2-ylethenyl)-1~{H}-quinoxalin-2-one, GLYCEROL, Pimtide, ... | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2023-10-31 | | Release date: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Pim1 in complex with (E)-3-(2-(thiophen-2-yl)vinyl)-3,4-dihydroquinoxalin-2(1H)-one and Pimtide
To Be Published
|
|
2KTZ
 
 | | Inhibitor Induced Structural Change in the HCV IRES Domain IIa RNA | | Descriptor: | (7R)-7-[(dimethylamino)methyl]-1-[3-(dimethylamino)propyl]-7,8-dihydro-1H-furo[3,2-e]benzimidazol-2-amine, HCV IRES Domain IIa RNA | | Authors: | Paulsen, R.B, Seth, P.P, Swayze, E.E, Griffey, R.H, Skalicky, J.J, Cheatham III, T.E, Davis, D.R. | | Deposit date: | 2010-02-10 | | Release date: | 2010-04-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Inhibitor-induced structural change in the HCV IRES domain IIa RNA.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2L62
 
 | | Protein and metal cluster structure of the wheat metallothionein domain g-Ec-1. The second part of the puzzle. | | Descriptor: | EC protein I/II, ZINC ION | | Authors: | Loebus, J, Peroza, E.A, Bluethgen, N, Fox, T, Meyer-Klaucke, W, Zerbe, O, Freisinger, E. | | Deposit date: | 2010-11-12 | | Release date: | 2011-05-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Protein and metal cluster structure of the wheat metallothionein domain gamma-E(c)-1: the second part of the puzzle.
J.Biol.Inorg.Chem., 16, 2011
|
|
6UL5
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with 4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenoxy}thieno[3,2-d]pyrimidin-2-yl)amino]-2-fluorobenzonitrile (24b), a non-nucleoside RT inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenoxy}thieno[3,2-d]pyrimidin-2-yl)amino]-2-fluorobenzonitrile, MAGNESIUM ION, ... | | Authors: | Ruiz, F.X, Pilch, A, Arnold, E. | | Deposit date: | 2019-10-06 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Discovery and Characterization of Fluorine-Substituted Diarylpyrimidine Derivatives as Novel HIV-1 NNRTIs with Highly Improved Resistance Profiles and Low Activity for the hERG Ion Channel.
J.Med.Chem., 63, 2020
|
|
3DUT
 
 | | The high salt (phosphate) crystal structure of deoxy hemoglobin E (GLU26LYS) at physiological pH (pH 7.35) | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PHOSPHATE ION, ... | | Authors: | Malashkevich, V.N, Balazs, T.C, Almo, S.C, Hirsch, R.E. | | Deposit date: | 2008-07-17 | | Release date: | 2009-08-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The high salt (phosphate) crystal structure of deoxy
hemoglobin E (GLU26LYS) at physiological pH (pH 7.35)
To be Published
|
|
6N57
 
 | | Cryo-EM structure of Escherichia coli RNAP polymerase bound with TraR in conformation I | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | Deposit date: | 2018-11-21 | | Release date: | 2020-02-26 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | E. coliTraR allosterically regulates transcription initiation by altering RNA polymerase conformation.
Elife, 8, 2019
|
|
6UTO
 
 | | Native E. coli Glyceraldehyde 3-phosphate dehydrogenase | | Descriptor: | ACETATE ION, Glyceraldehyde-3-phosphate dehydrogenase, SN-GLYCEROL-3-PHOSPHATE, ... | | Authors: | Rodriguez-Hernandez, A, Romo-Arevalo, E, Rodriguez-Romero, A. | | Deposit date: | 2019-10-29 | | Release date: | 2019-12-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A Novel Substrate-Binding Site in the X-Ray Structure of an Oxidized E. coli Glyceraldehyde 3-Phosphate Dehydrogenase Elucidated by Single-Wavelength Anomalous Dispersion
Crystals, 9, 2019
|
|
6NUP
 
 | |
6N58
 
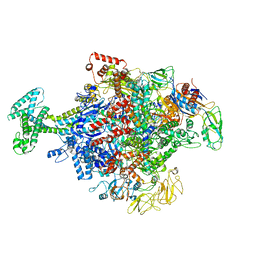 | | Cryo-EM structure of Escherichia coli RNAP polymerase bound with TraR in conformation II | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | Deposit date: | 2018-11-21 | | Release date: | 2020-02-26 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | E. coliTraR allosterically regulates transcription initiation by altering RNA polymerase conformation.
Elife, 8, 2019
|
|
5HR1
 
 | | Crystal structure of thioredoxin L107A mutant | | Descriptor: | COPPER (II) ION, Thioredoxin-1 | | Authors: | Noguera, M.E, Vazquez, D.S, Howard, E.I, Cousido-Siah, A, Mitschler, A, Podjarny, A, Santos, J. | | Deposit date: | 2016-01-22 | | Release date: | 2017-02-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.144 Å) | | Cite: | Structural variability of E. coli thioredoxin captured in the crystal structures of single-point mutants.
Sci Rep, 7, 2017
|
|
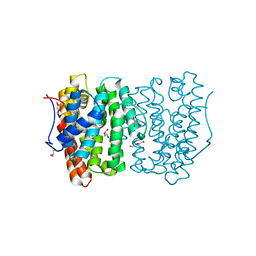
6OH7
 
 | |
1BP8
 
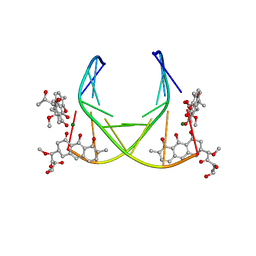 | | 4:2:1 mithramycin:Mg++:d(ACCCGGGT)2 complex | | Descriptor: | 1,2-HYDRO-1-OXY-3,4-HYDRO-3-(1-METHOXY-2-OXY-3,4-DIHYDROXYPENTYL)-8,9-DIHYDROXY-7-METHYL-ANTHRACENE, 2,6-dideoxy-3-C-methyl-beta-D-ribo-hexopyranose-(1-3)-2,6-dideoxy-beta-D-galactopyranose-(1-3)-beta-D-Olivopyranose, 5'-D(*AP*CP*CP*CP*GP*GP*GP*T)-3', ... | | Authors: | Keniry, M.A, Owen, E.A, Shafer, R.H. | | Deposit date: | 1998-08-13 | | Release date: | 1999-08-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of the 4:1 mithramycin:d(ACCCGGGT)2 complex: evidence for an interaction between the E saccharides
Biopolymers, 54, 2000
|
|
2LVF
 
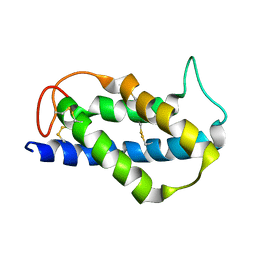 | | Solution structure of the Brazil Nut 2S albumin Ber e 1 | | Descriptor: | 2S albumin | | Authors: | Rundqvist, L, Tengel, T, Zdunek, J, Schleucher, J, Alcocer, M.J, Larsson, G. | | Deposit date: | 2012-07-04 | | Release date: | 2012-10-17 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure, copper binding and backbone dynamics of recombinant Ber e 1-the major allergen from Brazil nut.
Plos One, 7, 2012
|
|
2KCE
 
 | |
