6AYO
 
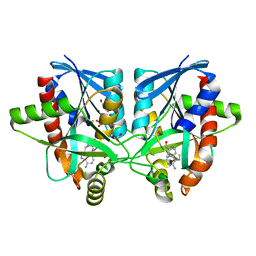 | | Crystal structure of Campylobacter jejuni 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with 5'-deoxy-5'-Propyl-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-propylpyrrolidin-3-ol, 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-09-08 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Transition-State Analogues of Campylobacter jejuni 5'-Methylthioadenosine Nucleosidase.
ACS Chem. Biol., 13, 2018
|
|
6AYM
 
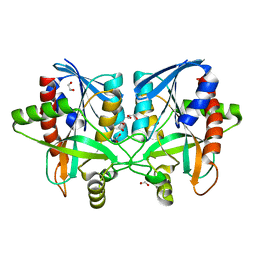 | | Crystal structure of Campylobacter jejuni 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) | | Descriptor: | 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | | Authors: | Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-09-08 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Transition-State Analogues of Campylobacter jejuni 5'-Methylthioadenosine Nucleosidase.
ACS Chem. Biol., 13, 2018
|
|
6AYS
 
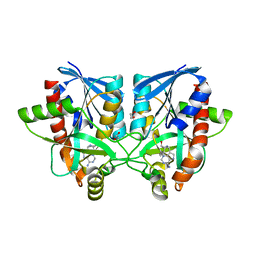 | | Crystal structure of Campylobacter jejuni 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with hexylthio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(hexylsulfanyl)methyl]pyrrolidin-3-ol, 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | | Authors: | Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-09-08 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Transition-State Analogues of Campylobacter jejuni 5'-Methylthioadenosine Nucleosidase.
ACS Chem. Biol., 13, 2018
|
|
6AYT
 
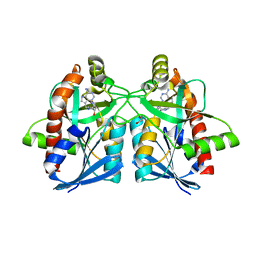 | | Crystal structure of Campylobacter jejuni 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with pyrazinylthio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(pyrazin-2-ylsulfanyl)methyl]pyrrolidin-3-ol, 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | | Authors: | Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-09-08 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Transition-State Analogues of Campylobacter jejuni 5'-Methylthioadenosine Nucleosidase.
ACS Chem. Biol., 13, 2018
|
|
8CPP
 
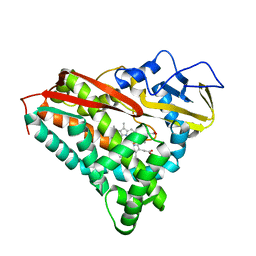 | | CRYSTAL STRUCTURES OF CYTOCHROME P450-CAM COMPLEXED WITH CAMPHANE, THIOCAMPHOR, AND ADAMANTANE: FACTORS CONTROLLING P450 SUBSTRATE HYDROXYLATION | | Descriptor: | CYTOCHROME P450CAM, PROTOPORPHYRIN IX CONTAINING FE, THIOCAMPHOR | | Authors: | Raag, R, Poulos, T.L. | | Deposit date: | 1990-05-18 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of cytochrome P-450CAM complexed with camphane, thiocamphor, and adamantane: factors controlling P-450 substrate hydroxylation.
Biochemistry, 30, 1991
|
|
5Z1Y
 
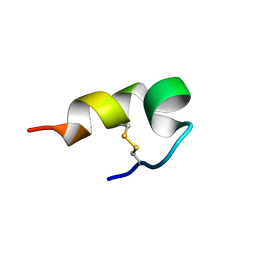 | | mBjAMP1 structure | | Descriptor: | mBjAMP1 peptide | | Authors: | Nam, J.Y, Lee, C.W. | | Deposit date: | 2017-12-28 | | Release date: | 2019-01-02 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | mBjAMP1 structure
To Be Published
|
|
6XU4
 
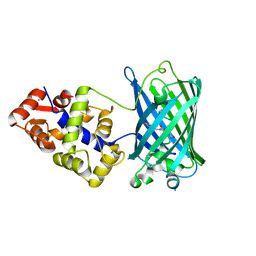 | | Crystal structure of the genetically-encoded FGCaMP calcium indicator in its calcium-bound state | | Descriptor: | CALCIUM ION, FGCamp | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Korzhenevskiy, D.A, Barykina, N.V, Subach, O.M, Subach, F.V. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | FGCaMP7, an Improved Version of Fungi-Based Ratiometric Calcium Indicator for In Vivo Visualization of Neuronal Activity.
Int J Mol Sci, 21, 2020
|
|
6XVP
 
 | | Crystal structure of Neprilysin in complex with Sampatrilat. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Neprilysin, ... | | Authors: | Cozier, G.E, Acharya, K.R, Sharma, U. | | Deposit date: | 2020-01-22 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Molecular Basis for Omapatrilat and Sampatrilat Binding to Neprilysin-Implications for Dual Inhibitor Design with Angiotensin-Converting Enzyme.
J.Med.Chem., 63, 2020
|
|
6X7Q
 
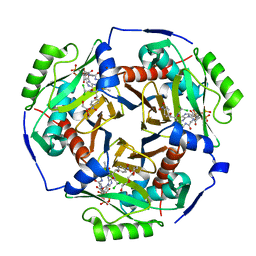 | | Chloramphenicol acetyltransferase type III in complex with chloramphenicol and acetyl-oxa(dethia)-CoA | | Descriptor: | CHLORAMPHENICOL, Chloramphenicol acetyltransferase 3, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Benjamin, A.B, Stunkard, L.M, Ling, J, Nice, J.N, Lohman, J.R. | | Deposit date: | 2020-05-30 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structures of chloramphenicol acetyltransferase III and Escherichia coli beta-keto-acylsynthase III co-crystallized with partially hydrolysed acetyl-oxa(de-thia)CoA
Acta Crystallogr.,Sect.F, 2023
|
|
6SVY
 
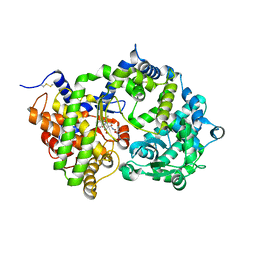 | | Crystal structure of Neprilysin in complex with Sampatrilat-ASP. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neprilysin, Sampatrilat-Asp, ... | | Authors: | Cozier, G.E, Acharya, K.R, Sharma, U. | | Deposit date: | 2019-09-19 | | Release date: | 2020-05-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Basis for Omapatrilat and Sampatrilat Binding to Neprilysin-Implications for Dual Inhibitor Design with Angiotensin-Converting Enzyme.
J.Med.Chem., 63, 2020
|
|
2Y21
 
 | |
3NU4
 
 | | Crystal Structure of HIV-1 Protease Mutant V32I with Antiviral Drug Amprenavir | | Descriptor: | CHLORIDE ION, SODIUM ION, protease, ... | | Authors: | Wang, Y.-F, Kovalevsky, A.Y, Weber, I.T. | | Deposit date: | 2010-07-06 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Amprenavir complexes with HIV-1 protease and its drug-resistant mutants altering hydrophobic clusters.
Febs J., 277, 2010
|
|
8OV5
 
 | | PERIDININ-CHLOROPHYLL-PROTEIN OF AMPHIDINIUM CARTERAE, 100K | | Descriptor: | CHLOROPHYLL A, PERIDININ, Peridinin-chlorophyll a-binding protein 1, ... | | Authors: | Hofmann, E, Johanning, S. | | Deposit date: | 2023-04-25 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural and spectroscopic characterization of the peridinin-chlorophyll a-protein (PCP) complex from Heterocapsa pygmaea (HPPCP).
Biochim Biophys Acta Bioenerg, 1866, 2024
|
|
7U2J
 
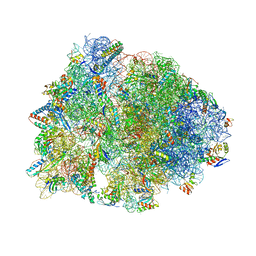 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with mRNA, aminoacylated A-site Gly-NH-tRNAgly, peptidyl P-site fMAC-NH-tRNAmet, deacylated E-site tRNAgly, and chloramphenicol at 2.55A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Syroegin, E.A, Aleksandrova, E.V, Polikanov, Y.S. | | Deposit date: | 2022-02-24 | | Release date: | 2022-07-13 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis for the inability of chloramphenicol to inhibit peptide bond formation in the presence of A-site glycine.
Nucleic Acids Res., 50, 2022
|
|
1NOO
 
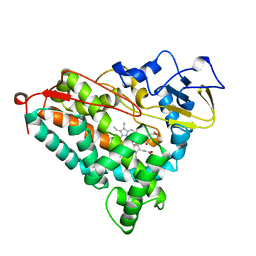 | | CYTOCHROME P450-CAM COMPLEXED WITH 5-EXO-HYDROXYCAMPHOR | | Descriptor: | 5-EXO-HYDROXYCAMPHOR, CYTOCHROME P450-CAM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, H.Y, Poulos, T.L. | | Deposit date: | 1995-12-02 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Cytochrome P450-Cam Complexed with its Catalytic Product, 5-Exo-Hydroxycamphor
J.Am.Chem.Soc., 117, 1995
|
|
3NU3
 
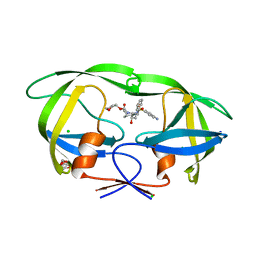 | | Wild Type HIV-1 Protease with Antiviral Drug Amprenavir | | Descriptor: | CHLORIDE ION, GLYCEROL, Protease, ... | | Authors: | Wang, Y.-F, Kovalevsky, A.Y, Weber, I.T. | | Deposit date: | 2010-07-06 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Amprenavir complexes with HIV-1 protease and its drug-resistant mutants altering hydrophobic clusters.
Febs J., 277, 2010
|
|
3NU6
 
 | |
3O2A
 
 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | GLUTAMIC ACID, GLYCEROL, Glutamate receptor 2, ... | | Authors: | Maclean, J.K.F, Basten, S, Campbell, R.A, Cumming, I.A, Gillen, K.J, Gillespie, J, Jamieson, C, Kazemier, B, Kiczun, M, Lamont, Y, Lyons, A.J, Moir, E.M, Morrow, J.A, Papakosta, M, Rankovic, Z, Smith, L. | | Deposit date: | 2010-07-22 | | Release date: | 2010-09-15 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel series of positive modulators of the AMPA receptor: discovery and structure based hit-to-lead studies.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
1SZO
 
 | | Crystal Structure Analysis of the 6-Oxo Camphor Hydrolase His122Ala Mutant Bound to Its Natural Product (2S,4S)-alpha-Campholinic Acid | | Descriptor: | (2S,4S)-4-(2,2-DIHYDROXYETHYL)-2,3,3-TRIMETHYLCYCLOPENTANONE, 6-oxocamphor hydrolase, CALCIUM ION | | Authors: | Leonard, P.M, Grogan, G. | | Deposit date: | 2004-04-06 | | Release date: | 2004-06-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of 6-oxo camphor hydrolase H122A mutant bound to its natural product, (2S,4S)-alpha-campholinic acid: mutant structure suggests an atypical mode of transition state binding for a crotonase homolog.
J.Biol.Chem., 279, 2004
|
|
3NU5
 
 | | Crystal Structure of HIV-1 Protease Mutant I50V with Antiviral Drug Amprenavir | | Descriptor: | ACETATE ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Wang, Y.-F, Shen, C.H, Weber, I.T. | | Deposit date: | 2010-07-06 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Amprenavir complexes with HIV-1 protease and its drug-resistant mutants altering hydrophobic clusters.
Febs J., 277, 2010
|
|
3O6I
 
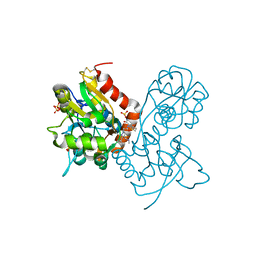 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | 2-[({3-tert-butyl-4-[(methylamino)methyl]-1H-pyrazol-1-yl}acetyl)amino]-4,5,6,7-tetrahydro-1-benzothiophene-3-carboxamide, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Maclean, J.K.F, Campbell, R.A, Cumming, I.A, Gillen, K.J, Gillespie, J, Jamieson, C, Kazemier, B, Kiczun, M, Lamont, Y, Lyons, A.J, Martin, F, Moir, E.M, Morrow, J.A, Pantling, J, Rankovic, Z, Smith, L. | | Deposit date: | 2010-07-29 | | Release date: | 2010-09-15 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel series of positive modulators of the AMPA receptor: structure-based lead optimization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3O29
 
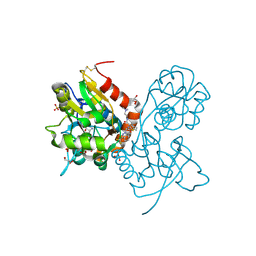 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | GLUTAMIC ACID, GLYCEROL, Glutamate receptor 2, ... | | Authors: | Maclean, J.K.F, Basten, S, Campbell, R.A, Cumming, I.A, Gillen, K.J, Gillespie, J, Jamieson, C, Kazemier, B, Kiczun, M, Lamont, Y, Lyons, A.J, Moir, E.M, Morrow, J.A, Papakosta, M, Rankovic, Z, Smith, L. | | Deposit date: | 2010-07-22 | | Release date: | 2010-09-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | A novel series of positive modulators of the AMPA receptor: discovery and structure based hit-to-lead studies.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3O28
 
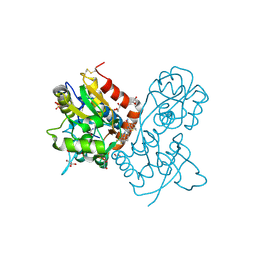 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | 2-({[3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazol-1-yl]acetyl}amino)-4,5,6,7-tetrahydro-1-benzothiophene-3-carboxamide, DIMETHYL SULFOXIDE, GLUTAMIC ACID, ... | | Authors: | Maclean, J.K.F, Basten, S, Campbell, R.A, Cumming, I.A, Gillen, K.J, Gillespie, J, Jamieson, C, Kazemier, B, Kiczun, M, Lamont, Y, Lyons, A.J, Moir, E.M, Morrow, J.A, Papakosta, M, Rankovic, Z, Smith, L. | | Deposit date: | 2010-07-22 | | Release date: | 2010-09-15 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel series of positive modulators of the AMPA receptor: discovery and structure based hit-to-lead studies.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3NUJ
 
 | |
2XAT
 
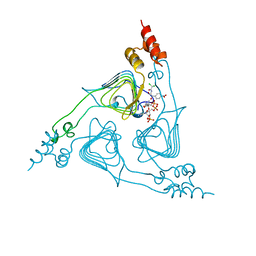 | |
