2VQC
 
 | | Structure of a DNA binding winged-helix protein, F-112, from Sulfolobus Spindle-shaped Virus 1. | | Descriptor: | HYPOTHETICAL 13.2 KDA PROTEIN | | Authors: | Menon, S.K, Kraft, P, Corn, G.J, Wiedenheft, B, Young, M.J, Lawrence, C.M. | | Deposit date: | 2008-03-12 | | Release date: | 2008-05-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cysteine Usage in Sulfolobus Spindle-Shaped Virus 1 and Extension to Hyperthermophilic Viruses in General.
Virology, 376, 2008
|
|
6QED
 
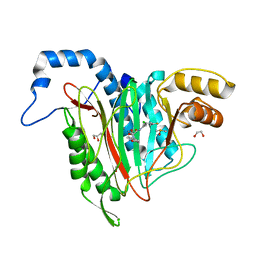 | | CRYSTAL STRUCTURE OF HUMAN METHIONINE AMINOPEPTIDASE-2 IN COMPLEX; WITH AN INHIBITOR (S)-3-Hydroxy-2-oxo-1-(2-oxo-1,2,3,4-tetrahydro-quinolin-6-yl)-pyrrolidine-3-carboxylic acid 3-chloro-5-fluoro-benzylamide | | Descriptor: | (3~{S})-~{N}-[(3-chloranyl-5-fluoranyl-phenyl)methyl]-3-oxidanyl-2-oxidanylidene-1-(2-oxidanylidene-3,4-dihydro-1~{H}-quinolin-6-yl)pyrrolidine-3-carboxamide, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Musil, D, Heinrich, T, Lehmann, M. | | Deposit date: | 2019-01-07 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery and Structure-Based Optimization of Next-Generation Reversible Methionine Aminopeptidase-2 (MetAP-2) Inhibitors.
J.Med.Chem., 62, 2019
|
|
4JF1
 
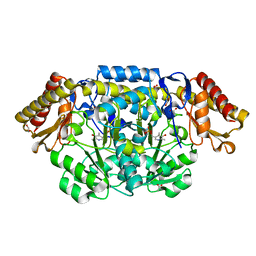 | | R144Q mutant of N-acetylornithine aminotransferase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Acetylornithine/succinyldiaminopimelate aminotransferase, ... | | Authors: | Bisht, S, Bharath, S.R, Murthy, M.R.N. | | Deposit date: | 2013-02-27 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Conformational transitions, ligand specificity and catalysis in N-acetylornithine aminotransferase: Implications on drug designing and rational enzyme engineering in omega aminotransferases
To be Published
|
|
5D7W
 
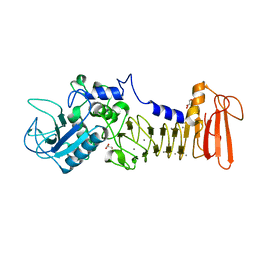 | | Crystal structure of serralysin | | Descriptor: | CALCIUM ION, GLYCEROL, Serralysin, ... | | Authors: | Wu, D, Ran, T, Xu, D.Q, Wang, W. | | Deposit date: | 2015-08-14 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure of a thermostable serralysin from Serratia sp. FS14 at 1.1 angstrom resolution.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
8KCU
 
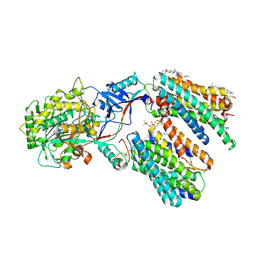 | | Cryo-EM structure of human gamma-secretase in complex with MK-0752 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Li, H, Kai, U, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2023-08-08 | | Release date: | 2024-08-14 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of human gamma-secretase inhibition by anticancer clinical compounds.
Nat.Struct.Mol.Biol., 32, 2025
|
|
5B2R
 
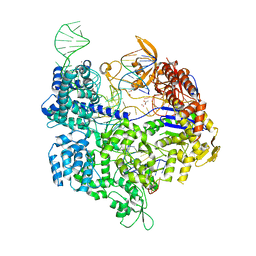 | | Crystal structure of the Streptococcus pyogenes Cas9 VQR variant in complex with sgRNA and target DNA (TGA PAM) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CRISPR-associated endonuclease Cas9, ... | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2016-02-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Altered PAM Specificities of Engineered CRISPR-Cas9
Mol.Cell, 61, 2016
|
|
3I2K
 
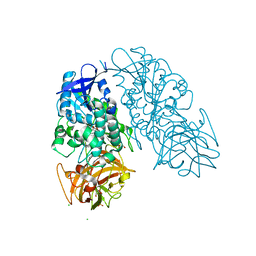 | | Cocaine Esterase, wild type, bound to a DTT adduct | | Descriptor: | (4S,5S)-4,5-BIS(MERCAPTOMETHYL)-1,3-DIOXOLAN-2-OL, CHLORIDE ION, Cocaine esterase, ... | | Authors: | Tesmer, J.J.G, Nance, M.R. | | Deposit date: | 2009-06-29 | | Release date: | 2010-06-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural analysis of thermostabilizing mutations of cocaine esterase.
Protein Eng.Des.Sel., 23, 2010
|
|
6CYD
 
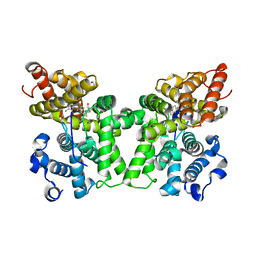 | | PDE2 in complex with compound 7 | | Descriptor: | 1,2-ETHANEDIOL, 3-(hydroxymethyl)-6-methyl-1-{(1S)-1-[4-(trifluoromethyl)phenyl]ethyl}-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, MAGNESIUM ION, ... | | Authors: | Lu, J. | | Deposit date: | 2018-04-05 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure-Guided Design and Procognitive Assessment of a Potent and Selective Phosphodiesterase 2A Inhibitor.
ACS Med Chem Lett, 9, 2018
|
|
1I7J
 
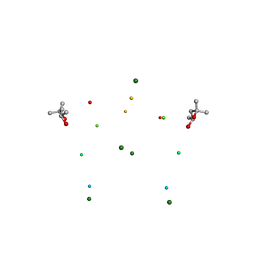 | | CRYSTAL STRUCTURE OF 2'-O-ME(CGCGCG)2: AN RNA DUPLEX AT 1.19 A RESOLUTION. 2-METHYL-2,4-PENTANEDIOL AND MAGNESIUM BINDING. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5'-R(*(OMC)P*(OMG)P*(OMC)P*(OMG)P*(OMC)P*(OMG))-3', MAGNESIUM ION | | Authors: | Adamiak, D.A, Rypniewski, W.R, Milecki, J, Adamiak, R.W. | | Deposit date: | 2001-03-09 | | Release date: | 2001-09-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | The 1.19 A X-ray structure of 2'-O-Me(CGCGCG)(2) duplex shows dehydrated RNA with 2-methyl-2,4-pentanediol in the minor groove.
Nucleic Acids Res., 29, 2001
|
|
6CZ7
 
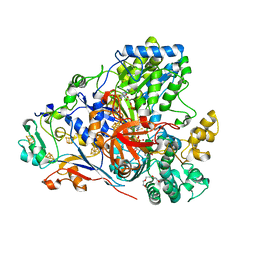 | | The arsenate respiratory reductase (Arr) complex from Shewanella sp. ANA-3 | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, 4Fe-4S ferredoxin, ... | | Authors: | Glasser, N.R, Newman, D.K. | | Deposit date: | 2018-04-08 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural and mechanistic analysis of the arsenate respiratory reductase provides insight into environmental arsenic transformations.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3HVO
 
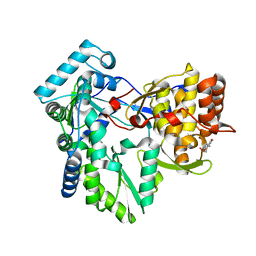 | | Structure of the genotype 2B HCV polymerase bound to a NNI | | Descriptor: | 2-(3-bromophenyl)-6-[(2-hydroxyethyl)amino]-1h-benzo[de]isoquinoline-1,3(2h)-dione, Genome polyprotein | | Authors: | Rydberg, E.H, Carfi, A. | | Deposit date: | 2009-06-16 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification and Biological Evaluation of a Series of 1H-Benzo[de]isoquinoline-1,3(2H)-diones as Hepatitis C Virus NS5B Polymerase Inhibitors
To be Published
|
|
6SA4
 
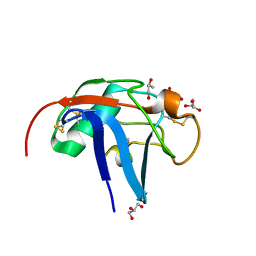 | | SALSA / DMBT1 / GP340 SRCR domain 1 | | Descriptor: | CHLORIDE ION, Deleted in malignant brain tumors 1 protein, GLYCEROL, ... | | Authors: | Reichhardt, M.P, Lea, S.M, Johnson, S. | | Deposit date: | 2019-07-16 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structures of SALSA/DMBT1 SRCR domains reveal the conserved ligand-binding mechanism of the ancient SRCR fold.
Life Sci Alliance, 3, 2020
|
|
6VTU
 
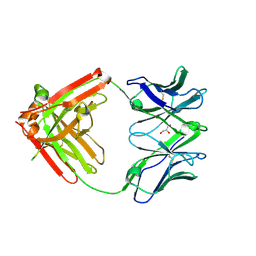 | | DH717.1 Fab monomer in complex with man9 glycan | | Descriptor: | DH717.1 heavy chain, DH717.1 light chain, GLYCEROL, ... | | Authors: | Fera, D, Bronkema, N. | | Deposit date: | 2020-02-13 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies.
Cell, 184, 2021
|
|
5ZRL
 
 | | M. smegmatis antimutator protein MutT2 in complex with CDP | | Descriptor: | 1,2-ETHANEDIOL, CYTIDINE-5'-DIPHOSPHATE, Putative mutator protein MutT2/NUDIX hydrolase | | Authors: | Singh, A, Arif, S.M, Sang, P.B, Varshney, U, Vijayan, M. | | Deposit date: | 2018-04-24 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural insights into the specificity and catalytic mechanism of mycobacterial nucleotide pool sanitizing enzyme MutT2.
J.Struct.Biol., 204, 2018
|
|
5BVA
 
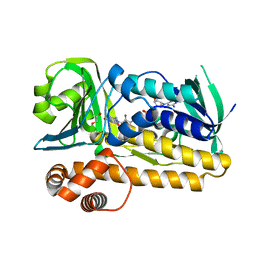 | | Structure of flavin-dependent brominase Bmp2 | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, flavin-dependent halogenase | | Authors: | Agarwal, V, Louie, G.V, Noel, J.P, Moore, B.S. | | Deposit date: | 2015-06-04 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.873 Å) | | Cite: | Biosynthesis of coral settlement cue tetrabromopyrrole in marine bacteria by a uniquely adapted brominase-thioesterase enzyme pair.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5BVK
 
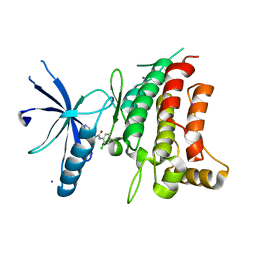 | | Fragment-based discovery of potent and selective DDR1/2 inhibitors | | Descriptor: | 1-(2-chlorophenyl)-3-(pyridin-3-ylmethyl)urea, Epithelial discoidin domain-containing receptor 1, IODIDE ION | | Authors: | Murray, C, Berdini, V, Buck, I, Carr, M, Cleasby, A, Coyle, J, Curry, J, Day, J, Hearn, K, Iqbal, A, Lee, L, Martins, V, Mortenson, P, Munck, J, Page, L, Patel, S, Roomans, S, Kirsten, T, Saxty, G. | | Deposit date: | 2015-06-05 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Fragment-Based Discovery of Potent and Selective DDR1/2 Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
5C0E
 
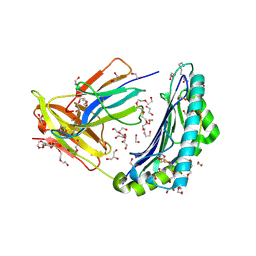 | | HLA-A02 carrying YLGGPDFPTI | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, Beta-2-microglobulin, ... | | Authors: | Rizkallah, P.J, Bulek, A.M, Cole, D.K, Sewell, A.K. | | Deposit date: | 2015-06-12 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.491 Å) | | Cite: | Hotspot autoimmune T cell receptor binding underlies pathogen and insulin peptide cross-reactivity.
J.Clin.Invest., 126, 2016
|
|
5AP7
 
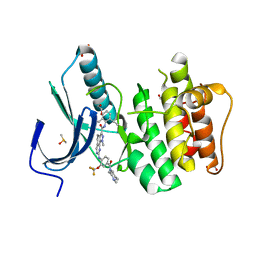 | | Naturally Occurring Mutations in the MPS1 Gene Predispose Cells to Kinase Inhibitor Drug Resistance. | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, MONOPOLAR SPINDLE KINASE 1, ... | | Authors: | Gurden, M.D, Westwood, I.M, Faisal, A, Naud, S, Cheung, K.M, McAndrew, C, Wood, A, Schmitt, J, Boxall, K, Mak, G, Workman, P, Burke, R, Hoelder, S, Blagg, J, van Montfort, R, Linardopoulos, S. | | Deposit date: | 2015-09-14 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Naturally Occurring Mutations in the Mps1 Gene Predispose Cells to Kinase Inhibitor Drug Resistance.
Cancer Res., 75, 2015
|
|
5C5D
 
 | |
3NPF
 
 | |
5P8Y
 
 | | humanized rat catechol O-methyltransferase in complex with 5-(4-fluorophenyl)-2,3-dihydroxy-N-[3-(pyrrolo[3,2-c]pyridin-1-ylmethoxy)propyl]benzamide at 1.42A | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 5-(4-fluorophenyl)-2,3-dihydroxy-N-[3-(pyrrolo[3,2-c]pyridin-1-ylmethoxy)propyl]benzamide, ... | | Authors: | Ehler, A, Lerner, C, Rudolph, M.G. | | Deposit date: | 2016-08-29 | | Release date: | 2017-08-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal Structure of a COMT complex
To be published
|
|
5Y9Z
 
 | | Crystal structure of rat hematopoietic prostaglandin D synthase | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, GLUTATHIONE, ... | | Authors: | Kamo, M, Furubayashi, N, Inaka, K, Tanaka, H, Aritake, K, Urade, Y. | | Deposit date: | 2017-08-29 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Crystal structure of rat hematopoietic prostaglandin D synthase
To Be Published
|
|
3L17
 
 | | Discovery of (thienopyrimidin-2-yl)aminopyrimidines as Potent, Selective, and Orally Available Pan-PI3-Kinase and Dual Pan-PI3-Kinase/mTOR Inhibitors for the Treatment of Cancer | | Descriptor: | 4-methyl-5-(6-{[4-(methylsulfonyl)piperazin-1-yl]methyl}-4-morpholin-4-ylthieno[3,2-d]pyrimidin-2-yl)pyrimidin-2-amine, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Murray, J.M, Wiesmann, C. | | Deposit date: | 2009-12-10 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of (Thienopyrimidin-2-yl)aminopyrimidines as Potent, Selective, and Orally Available Pan-PI3-Kinase and Dual Pan-PI3-Kinase/mTOR Inhibitors for the Treatment of Cancer.
J.Med.Chem., 53, 2010
|
|
7VT7
 
 | | Crystal structure of CBM deleted MtGlu5 in complex with CBI | | Descriptor: | Endoglucanase H, GLYCEROL, TRYPTOPHAN, ... | | Authors: | Ye, T.J, Ko, P.T, Huang, K.F, Wu, S.H. | | Deposit date: | 2021-10-28 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Synergic action of an inserted carbohydrate-binding module in a glycoside hydrolase family 5 endoglucanase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
3C7R
 
 | | Crystal Structure of HIV-1 subtype F DIS extended duplex RNA bound to neomycin | | Descriptor: | HIV-1 subtype F genomic RNA, NEOMYCIN, POTASSIUM ION | | Authors: | Freisz, S, Lang, K, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-02-08 | | Release date: | 2008-05-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Binding of aminoglycoside antibiotics to the duplex form of the HIV-1 genomic RNA dimerization initiation site.
Angew.Chem.Int.Ed.Engl., 47, 2008
|
|
