1CDK
 
 | | CAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT (E.C.2.7.1.37) (PROTEIN KINASE A) COMPLEXED WITH PROTEIN KINASE INHIBITOR PEPTIDE FRAGMENT 5-24 (PKI(5-24) ISOELECTRIC VARIANT CA) AND MN2+ ADENYLYL IMIDODIPHOSPHATE (MNAMP-PNP) AT PH 5.6 AND 7C AND 4C | | Descriptor: | CAMP-DEPENDENT PROTEIN KINASE, MANGANESE (II) ION, MYRISTIC ACID, ... | | Authors: | Bossemeyer, D, Engh, R.A, Kinzel, V, Ponstingl, H, Huber, R. | | Deposit date: | 1994-07-04 | | Release date: | 1995-10-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphotransferase and substrate binding mechanism of the cAMP-dependent protein kinase catalytic subunit from porcine heart as deduced from the 2.0 A structure of the complex with Mn2+ adenylyl imidodiphosphate and inhibitor peptide PKI(5-24).
EMBO J., 12, 1993
|
|
1BTI
 
 | | CREVICE-FORMING MUTANTS IN THE RIGID CORE OF BOVINE PANCREATIC TRYPSIN INHIBITOR: CRYSTAL STRUCTURES OF F22A, Y23A, N43G, AND F45A | | Descriptor: | BOVINE PANCREATIC TRYPSIN INHIBITOR | | Authors: | Housset, D, Tao, F, Kim, K.-S, Fuchs, J, Woodward, C, Wlodawer, A. | | Deposit date: | 1991-07-11 | | Release date: | 1993-10-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crevice-forming mutants in the rigid core of bovine pancreatic trypsin inhibitor: crystal structures of F22A, Y23A, N43G, and F45A.
Protein Sci., 2, 1993
|
|
1BJN
 
 | |
1BJW
 
 | | ASPARTATE AMINOTRANSFERASE FROM THERMUS THERMOPHILUS | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PHOSPHATE ION | | Authors: | Nakai, T, Okada, K, Kuramitsu, S, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1998-06-30 | | Release date: | 1999-07-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Thermus thermophilus HB8 aspartate aminotransferase and its complex with maleate.
Biochemistry, 38, 1999
|
|
1BSR
 
 | |
1CE6
 
 | | MHC CLASS I H-2DB COMPLEXED WITH A SENDAI VIRUS NUCLEOPROTEIN PEPTIDE | | Descriptor: | PROTEIN (HUMAN BETA-2 MICROGLOBULIN), PROTEIN (MHC CLASS I H-2DB HEAVY CHAIN), PROTEIN (SENDAI VIRUS NUCLEOPROTEIN), ... | | Authors: | Tormo, J, Jones, E.Y. | | Deposit date: | 1999-03-17 | | Release date: | 1999-03-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of two H-2Db/glycopeptide complexes suggest a molecular basis for CTL cross-reactivity.
Immunity, 10, 1999
|
|
1CHZ
 
 | | A NEW NEUROTOXIN FROM BUTHUS MARTENSII KARSCH | | Descriptor: | CHLORIDE ION, PROTEIN (BMK M2) | | Authors: | He, X.L, Deng, J.P, Li, H.M, Wang, D.C. | | Deposit date: | 1999-03-31 | | Release date: | 2000-03-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure of a new neurotoxin from the scorpion Buthus martensii Karsch at 1.76 A.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1CI6
 
 | | TRANSCRIPTION FACTOR ATF4-C/EBP BETA BZIP HETERODIMER | | Descriptor: | BETA-MERCAPTOETHANOL, FE (III) ION, TRANSCRIPTION FACTOR ATF-4, ... | | Authors: | Podust, L.M, Kim, Y. | | Deposit date: | 1999-04-07 | | Release date: | 2000-12-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the CCAAT box/enhancer-binding protein beta activating transcription factor-4 basic leucine zipper heterodimer in the absence of DNA
J.Biol.Chem., 276, 2001
|
|
1CL7
 
 | | ANTI HIV1 PROTEASE FAB | | Descriptor: | PROTEIN (IGG1 ANTIBODY 1696 (constant heavy chain)), PROTEIN (IGG1 ANTIBODY 1696 (light chain)), PROTEIN (IGG1 ANTIBODY 1696 (variable heavy chain)) | | Authors: | Lescar, J, Bentley, G.A. | | Deposit date: | 1999-05-06 | | Release date: | 2000-01-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Inhibition of the HIV-1 and HIV-2 proteases by a monoclonal antibody.
Protein Sci., 8, 1999
|
|
1CLK
 
 | | CRYSTAL STRUCTURE OF STREPTOMYCES DIASTATICUS NO.7 STRAIN M1033 XYLOSE ISOMERASE AT 1.9 A RESOLUTION WITH PSEUDO-I222 SPACE GROUP | | Descriptor: | COBALT (II) ION, MAGNESIUM ION, XYLOSE ISOMERASE | | Authors: | Niu, L, Teng, M, Zhu, X, Gong, W. | | Deposit date: | 1999-04-29 | | Release date: | 2000-05-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of xylose isomerase from Streptomyces diastaticus no. 7 strain M1033 at 1.85 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1CLW
 
 | | TAILSPIKE PROTEIN FROM PHAGE P22, V331A MUTANT | | Descriptor: | TAILSPIKE PROTEIN | | Authors: | Steinbacher, S, Baxa, U, Weintraub, A, Huber, R, Seckler, R. | | Deposit date: | 1999-05-04 | | Release date: | 1999-11-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutations improving the folding of phage P22 tailspike protein affect its receptor binding activity.
J.Mol.Biol., 293, 1999
|
|
1CVX
 
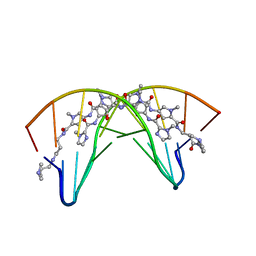 | | CRYSTAL STRUCTURE OF POLYAMIDE DIMER (IMPYHPPYBETADP)2 BOUND TO B-DNA DECAMER CCAGATCTGG | | Descriptor: | 5'-D(*CP*CP*AP*GP*AP*TP*CP*TP*GP*G)-3', HYDROXYPYRROLE-IMIDAZOLE-PYRROLE POLYAMIDE | | Authors: | Kielkopf, C.L, Bremer, R.E, White, S, Baird, E.E, Dervan, P.B, Rees, D.C. | | Deposit date: | 1999-08-24 | | Release date: | 2000-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural effects of DNA sequence on T.A recognition by hydroxypyrrole/pyrrole pairs in the minor groove.
J.Mol.Biol., 295, 2000
|
|
1CW4
 
 | |
1CWH
 
 | | HUMAN CYCLOPHILIN A COMPLEXED WITH 3-D-SER CYCLOSPORIN | | Descriptor: | CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | Authors: | Mikol, V, Kallen, J, Taylor, P, Walkinshaw, M.D. | | Deposit date: | 1998-05-07 | | Release date: | 1998-07-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | X-Ray Structures and Analysis of 11 Cyclosporin Derivatives Complexed with Cyclophilin A.
J.Mol.Biol., 283, 1998
|
|
1CET
 
 | | CHLOROQUINE BINDS IN THE COFACTOR BINDING SITE OF PLASMODIUM FALCIPARUM LACTATE DEHYDROGENASE. | | Descriptor: | N4-(7-CHLORO-QUINOLIN-4-YL)-N1,N1-DIETHYL-PENTANE-1,4-DIAMINE, PROTEIN (L-LACTATE DEHYDROGENASE) | | Authors: | Read, J.A, Wilkinson, K.W, Tranter, R, Sessions, R.B, Brady, R.L. | | Deposit date: | 1999-03-10 | | Release date: | 1999-03-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Chloroquine binds in the cofactor binding site of Plasmodium falciparum lactate dehydrogenase.
J.Biol.Chem., 274, 1999
|
|
1CF5
 
 | | BETA-MOMORCHARIN STRUCTURE AT 2.55 A | | Descriptor: | PROTEIN (BETA-MOMORCHARIN), beta-D-xylopyranose-(1-2)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yuan, Y.-R, He, Y.-N, Xiong, J.-P, Xia, Z.-X. | | Deposit date: | 1999-03-24 | | Release date: | 1999-06-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Three-dimensional structure of beta-momorcharin at 2.55 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1CIY
 
 | |
1CZ3
 
 | | DIHYDROFOLATE REDUCTASE FROM THERMOTOGA MARITIMA | | Descriptor: | DIHYDROFOLATE REDUCTASE, SULFATE ION | | Authors: | Dams, T, Auerbach, G, Bader, G, Ploom, T, Huber, R, Jaenicke, R. | | Deposit date: | 1999-09-01 | | Release date: | 2000-03-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of dihydrofolate reductase from Thermotoga maritima: molecular features of thermostability.
J.Mol.Biol., 297, 2000
|
|
1CJV
 
 | | COMPLEX OF GS-ALPHA WITH THE CATALYTIC DOMAINS OF MAMMALIAN ADENYLYL CYCLASE: COMPLEX WITH BETA-L-2',3'-DIDEOXYATP, MG, AND ZN | | Descriptor: | 2',3'-DIDEOXYADENOSINE-5'-TRIPHOSPHATE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ... | | Authors: | Tesmer, J.J.G, Sprang, S.R. | | Deposit date: | 1999-04-19 | | Release date: | 1999-08-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Two-metal-Ion catalysis in adenylyl cyclase.
Science, 285, 1999
|
|
1CKE
 
 | | CMP KINASE FROM ESCHERICHIA COLI FREE ENZYME STRUCTURE | | Descriptor: | PROTEIN (CYTIDINE MONOPHOSPHATE KINASE), SULFATE ION | | Authors: | Briozzo, P, Golinelli-Pimpaneau, B. | | Deposit date: | 1998-09-24 | | Release date: | 1999-09-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of escherichia coli CMP kinase alone and in complex with CDP: a new fold of the nucleoside monophosphate binding domain and insights into cytosine nucleotide specificity.
Structure, 6, 1998
|
|
1D1G
 
 | | DIHYDROFOLATE REDUCTASE FROM THERMOTOGA MARITIMA | | Descriptor: | DIHYDROFOLATE REDUCTASE, METHOTREXATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Dams, T, Auerbach, G, Bader, G, Ploom, T, Huber, R, Jaenicke, R. | | Deposit date: | 1999-09-16 | | Release date: | 2000-03-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of dihydrofolate reductase from Thermotoga maritima: molecular features of thermostability.
J.Mol.Biol., 297, 2000
|
|
1CLI
 
 | | X-RAY CRYSTAL STRUCTURE OF AMINOIMIDAZOLE RIBONUCLEOTIDE SYNTHETASE (PURM), FROM THE E. COLI PURINE BIOSYNTHETIC PATHWAY, AT 2.5 A RESOLUTION | | Descriptor: | PROTEIN (PHOSPHORIBOSYL-AMINOIMIDAZOLE SYNTHETASE), SULFATE ION | | Authors: | Li, C, Kappock, T.J, Stubbe, J, Weaver, T.M, Ealick, S.E. | | Deposit date: | 1999-04-28 | | Release date: | 1999-10-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray crystal structure of aminoimidazole ribonucleotide synthetase (PurM), from the Escherichia coli purine biosynthetic pathway at 2.5 A resolution.
Structure Fold.Des., 7, 1999
|
|
1CM7
 
 | | 3-ISOPROPYLMALATE DEHYDROGENASE FROM ESCHERICHIA COLI | | Descriptor: | PROTEIN (3-ISOPROPYLMALATE DEHYDROGENASE) | | Authors: | Wallon, G, Kryger, G, Lovett, S.T, Oshima, T, Ringe, D, Petsko, G.A. | | Deposit date: | 1999-05-17 | | Release date: | 1999-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structures of Escherichia coli and Salmonella typhimurium 3-isopropylmalate dehydrogenase and comparison with their thermophilic counterpart from Thermus thermophilus.
J.Mol.Biol., 266, 1997
|
|
1D3V
 
 | | CRYSTAL STRUCTURE OF THE BINUCLEAR MANGANESE METALLOENZYME ARGINASE COMPLEXED WITH 2(S)-AMINO-6-BORONOHEXANOIC ACID, AN L-ARGININE ANALOG | | Descriptor: | 2(S)-AMINO-6-BORONOHEXANOIC ACID, MANGANESE (II) ION, PROTEIN (ARGINASE) | | Authors: | Cox, J.D, Kim, N.N, Traish, A.M, Christianson, D.W. | | Deposit date: | 1999-10-01 | | Release date: | 1999-11-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Arginase-boronic acid complex highlights a physiological role in erectile function.
Nat.Struct.Biol., 6, 1999
|
|
1D4M
 
 | | THE CRYSTAL STRUCTURE OF COXSACKIEVIRUS A9 TO 2.9 A RESOLUTION | | Descriptor: | 5-(7-(4-(4,5-DIHYDRO-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, MYRISTIC ACID, PROTEIN (COXSACKIEVIRUS A9) | | Authors: | Hendry, E, Hatanaka, H, Fry, E, Smyth, M, Tate, J, Stanway, G, Santti, J, Maaronen, M, Hyypia, T, Stuart, D. | | Deposit date: | 1999-10-04 | | Release date: | 1999-12-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of coxsackievirus A9: new insights into the uncoating mechanisms of enteroviruses.
Structure Fold.Des., 7, 1999
|
|
