7AAG
 
 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-617 | | Descriptor: | 1,2-ETHANEDIOL, 1-cycloheptyl-3-(4-methoxy-3-{4-[4-(1H-1,2,3,4-tetrazol-5-yl)phenoxy]butoxy}phenyl)-4,4-dimethyl-4,5-dihydro-1H-pyrazol-5-one, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Singh, A.K, Blaazer, A.R, Zara, L, de Esch, I.J.P, Leurs, R, Brown, D.G. | | Deposit date: | 2020-09-04 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | hPDE4D2 structure with inhibitor NPD-617
To be published
|
|
1RKX
 
 | | Crystal Structure at 1.8 Angstrom of CDP-D-glucose 4,6-dehydratase from Yersinia pseudotuberculosis | | Descriptor: | CDP-glucose-4,6-dehydratase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Vogan, E.M, Bellamacina, C, He, X, Liu, H.W, Ringe, D, Petsko, G.A. | | Deposit date: | 2003-11-23 | | Release date: | 2004-03-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure at 1.8 A Resolution of CDP-d-Glucose 4,6-Dehydratase from Yersinia pseudotuberculosis
Biochemistry, 43, 2004
|
|
4A9X
 
 | | Pseudomonas fluorescens PhoX in complex with the substrate analogue AppCp | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, MU-OXO-DIIRON, ... | | Authors: | Yong, S.C, Roversi, P, Lillington, J.E.D, Zeldin, O.B, Garman, E.F, Lea, S.M, Berks, B.C. | | Deposit date: | 2011-11-29 | | Release date: | 2012-12-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A Complex Iron-Calcium Cofactor Catalyzing Phosphotransfer Chemistry
Science, 345, 2014
|
|
3JZI
 
 | | Crystal structure of biotin carboxylase from E. Coli in complex with benzimidazole series | | Descriptor: | 7-amino-2-[(2-chlorobenzyl)amino]-1-{[(1S,2S)-2-hydroxycycloheptyl]methyl}-1H-benzimidazole-5-carboxamide, Biotin carboxylase | | Authors: | Cheng, C, Shipps, G.W, Yang, Z, Sun, B, Kawahata, N, Soucy, K, Soriano, A, Orth, P, Xiao, L, Mann, P, Black, T. | | Deposit date: | 2009-09-23 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Discovery and optimization of antibacterial AccC inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6UO6
 
 | |
1LIF
 
 | |
6L2Q
 
 | | Threonyl-tRNA synthetase from Salmonella enterica in complex with an inhibitor | | Descriptor: | (2S,3R)-2-azanyl-N-[(E)-4-[6,7-bis(chloranyl)-4-oxidanylidene-quinazolin-3-yl]but-2-enyl]-3-oxidanyl-butanamide, 1,2-ETHANEDIOL, Threonine--tRNA ligase, ... | | Authors: | Guo, J, Chen, B, Zhou, H. | | Deposit date: | 2019-10-05 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of novel tRNA-amino acid dual-site inhibitors against threonyl-tRNA synthetase by fragment-based target hopping.
Eur.J.Med.Chem., 187, 2019
|
|
4ZO5
 
 | | PDE10 complexed with 4-isopropoxy-2-(2-(3-(4-methoxyphenyl)-4-oxo-3,4-dihydroquinazolin-2-yl)ethyl)isoindoline-1,3-dione | | Descriptor: | 2-{2-[3-(4-methoxyphenyl)-4-oxo-3,4-dihydroquinazolin-2-yl]ethyl}-4-(propan-2-yloxy)-1H-isoindole-1,3(2H)-dione, 3-(1-hydroxy-2-methylpropan-2-yl)-5-phenyl-3,5-dihydro-1H-imidazo[4,5-c][1,8]naphthyridine-2,4-dione, MAGNESIUM ION, ... | | Authors: | Yan, Y. | | Deposit date: | 2015-05-06 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of [(11)C]MK-8193 as a PET tracer to measure target engagement of phosphodiesterase 10A (PDE10A) inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
6QRR
 
 | | X-ray radiation dose series on xylose isomerase - 0.13 MGy | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, MAGNESIUM ION, ... | | Authors: | Taberman, H, Bury, C.S, van der Woerd, M.J, Snell, E.H, Garman, E.F. | | Deposit date: | 2019-02-19 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.096 Å) | | Cite: | Structural knowledge or X-ray damage? A case study on xylose isomerase illustrating both.
J.Synchrotron Radiat., 26, 2019
|
|
2QJV
 
 | |
6QMK
 
 | | Small molecule inhibitor of the KEAP1-NRF2 protein-protein interaction | | Descriptor: | (3~{S})-3-[3-[[1,1-bis(oxidanylidene)-3,4-dihydro-5,1$l^{6},2-benzoxathiazepin-2-yl]methyl]-4-methyl-phenyl]-3-(7-methoxy-1-methyl-benzotriazol-5-yl)propanoic acid, CHLORIDE ION, Kelch-like ECH-associated protein 1 | | Authors: | Davies, T.G. | | Deposit date: | 2019-02-07 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structure-Activity and Structure-Conformation Relationships of Aryl Propionic Acid Inhibitors of the Kelch-like ECH-Associated Protein 1/Nuclear Factor Erythroid 2-Related Factor 2 (KEAP1/NRF2) Protein-Protein Interaction.
J.Med.Chem., 62, 2019
|
|
6EVA
 
 | | Structure of E277Q A. niger fdc1 in complex with a phenylpyruvate derived adduct to the prenylated flavin cofactor | | Descriptor: | 1-deoxy-5-O-phosphono-1-[(1S)-3,3,4,5-tetramethyl-9,11-dioxo-1-(phenylacetyl)-2,3,8,9,10,11-hexahydro-1H,7H-quinolino[1 ,8-fg]pteridin-7-yl]-D-ribitol, Ferulic acid decarboxylase 1, MANGANESE (II) ION, ... | | Authors: | Bailey, S.S, David, L, Payne, K.A.P. | | Deposit date: | 2017-11-01 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The role of conserved residues in Fdc decarboxylase in prenylated flavin mononucleotide oxidative maturation, cofactor isomerization, and catalysis.
J. Biol. Chem., 293, 2018
|
|
2QHA
 
 | |
6U0Q
 
 | | Transthyretin in complex with (E)-5,5'-(ethene-1,2-diyl)bis(1,1-dihydroxy-3-oxo-1,3-dihydrobenzo[c][1,2]oxaborol-1-uide) | | Descriptor: | 3-[(~{E})-2-[7,7-bis(oxidanyl)-9-oxidanylidene-8-oxa-7-boranuidabicyclo[4.3.0]nona-1,3,5-trien-3-yl]ethenyl]-7,7-bis(oxidanyl)-8-oxa-7-boranuidabicyclo[4.3.0]nona-1(6),2,4-trien-9-one, Transthyretin | | Authors: | Windsor, I.W, Graham, B.J, Raines, R.T. | | Deposit date: | 2019-08-14 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Boronic acid with high oxidative stability and utility in biological contexts.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4GRB
 
 | | Casein kinase 2 (CK2) bound to inhibitor | | Descriptor: | 5-(2-{[4-(dimethylcarbamoyl)phenyl]amino}-4-methoxypyrimidin-5-yl)thiophene-3-carboxylic acid, CHLORIDE ION, Casein kinase II subunit alpha | | Authors: | Larsen, N.A, Dowling, J.E, Ferguson, A.D. | | Deposit date: | 2012-08-24 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and Property Based Design of Pyrazolo[1,5-a]pyrimidine Inhibitors of CK2 Kinase with Activity in Vivo.
ACS Med Chem Lett, 4, 2013
|
|
2QT0
 
 | | Human nicotinamide riboside kinase 1 in complex with nicotinamide riboside and an ATP analogue | | Descriptor: | MAGNESIUM ION, Nicotinamide riboside, Nicotinamide riboside kinase 1, ... | | Authors: | Rabeh, W.M, Tempel, W, Nedyalkova, L, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Brenner, C, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-31 | | Release date: | 2007-08-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Nicotinamide Riboside Kinase Structures Reveal New Pathways to NAD(+).
Plos Biol., 5, 2007
|
|
5Y8Y
 
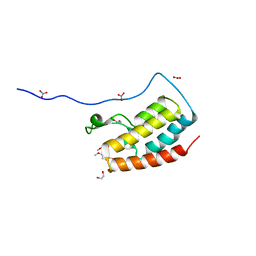 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromanyl-2-methoxy-N-(6-methoxy-3-methyl-1,2-benzoxazol-5-yl)benzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Song, M, Wang, C. | | Deposit date: | 2017-08-21 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
7FXC
 
 | | Crystal Structure of human FABP4 in complex with 6-[(3,4-dichlorophenyl)methyl]-5-methyl-1,1-dioxo-1,2,6-thiadiazin-3-one, i.e. SMILES N1(Cc2cc(c(cc2)Cl)Cl)S(=O)(=O)NC(=O)C=C1C with IC50=0.945 microM | | Descriptor: | 6-[(3,4-dichlorophenyl)methyl]-5-methyl-1lambda~6~,2,6-thiadiazine-1,1,3(2H,6H)-trione, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Albrecht-Harry, A, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
1LIB
 
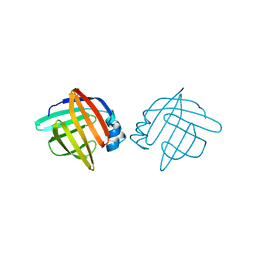 | |
7G1R
 
 | | Crystal Structure of human FABP4 in complex with 2-[(3-ethoxycarbonylthiophen-2-yl)carbamoyl]cyclopentene-1-carboxylic acid, i.e. SMILES C1(=C(C=CS1)C(=O)OCC)NC(=O)C1=C(C(=O)O)CCC1 with IC50=1.4 microM | | Descriptor: | 2-{[3-(ethoxycarbonyl)thiophen-2-yl]carbamoyl}cyclopent-1-ene-1-carboxylic acid, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Ceccarelli-Simona, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
7FX1
 
 | | Crystal Structure of human FABP4 in complex with 2-(3-phenyl-4-piperidin-1-ylphenyl)acetic acid, i.e. SMILES c1(c(N2CCCCC2)ccc(c1)CC(=O)O)c1ccccc1 with IC50=0.252291 microM | | Descriptor: | Fatty acid-binding protein, adipocyte, SULFATE ION, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Pfister, R, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
9CRF
 
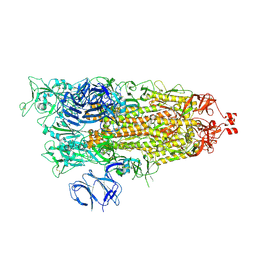 | | Cryo-EM structure of SARS-CoV-2 Spike Proteins on intact virions: Alpha (B.1.1.7) variant 1 open RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ke, Z, Croll, T.I, Briggs, J.A.G. | | Deposit date: | 2024-07-22 | | Release date: | 2024-11-27 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Virion morphology and on-virus spike protein structures of diverse SARS-CoV-2 variants.
Embo J., 43, 2024
|
|
6I9R
 
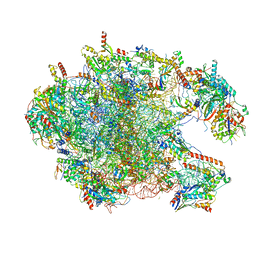 | | Large subunit of the human mitochondrial ribosome in complex with Virginiamycin M and Quinupristin | | Descriptor: | 16S rRNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Modelska, A, Aibara, S, Amunts, A. | | Deposit date: | 2018-11-25 | | Release date: | 2020-07-08 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Inhibition of mitochondrial translation suppresses glioblastoma stem cell growth.
Cell Rep, 35, 2021
|
|
2YL6
 
 | | Inhibition of the pneumococcal virulence factor StrH and molecular insights into N-glycan recognition and hydrolysis | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, BETA-N-ACETYLHEXOSAMINIDASE, ... | | Authors: | Pluvinage, B, Higgins, M.A, Abbott, D.W, Robb, C, Dalia, A.B, Deng, L, Weiser, J.N, Parsons, T.B, Fairbanks, A.J, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-05-31 | | Release date: | 2011-09-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Inhibition of the Pneumococcal Virulence Factor Strh and Molecular Insights Into N-Glycan Recognition and Hydrolysis.
Structure, 19, 2011
|
|
7G13
 
 | | Crystal Structure of human FABP4 binding site mutated to that of FABP5 in complex with 4-[[3-(3-cyclopropyl-1,2,4-oxadiazol-5-yl)-4,5,6,7-tetrahydro-1-benzothiophen-2-yl]carbamoyl]-3,6-dihydro-2H-pyran-5-carboxylic acid, i.e. SMILES S1C(=C(C2=NC(=NO2)C2CC2)C2=C1CCCC2)NC(=O)C1=C(COCC1)C(=O)O with IC50=0.119472 microM | | Descriptor: | 4-{[(3M)-3-(3-cyclopropyl-1,2,4-oxadiazol-5-yl)-4,5,6,7-tetrahydro-1-benzothiophen-2-yl]carbamoyl}-5,6-dihydro-2H-pyran-3-carboxylic acid, DIMETHYL SULFOXIDE, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Richter, H, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
