1RPN
 
 | | Crystal Structure of GDP-D-mannose 4,6-dehydratase in complexes with GDP and NADPH | | Descriptor: | GDP-mannose 4,6-dehydratase, GUANOSINE-5'-DIPHOSPHATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Webb, N.A, Mulichak, A.M, Lam, J.S, Rocchetta, H.L, Garavito, R.M. | | Deposit date: | 2003-12-03 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of a tetrameric GDP-D-mannose 4,6-dehydratase from a bacterial GDP-D-rhamnose biosynthetic pathway.
Protein Sci., 13, 2004
|
|
1PU2
 
 | | Crystal Structure of the K246R Mutant of Aspartate Semialdehyde Dehydrogenase from Haemophilus influenzae | | Descriptor: | Aspartate-semialdehyde dehydrogenase | | Authors: | Blanco, J, Moore, R.A, Faehnle, C.R, Coe, D.M, Viola, R.E. | | Deposit date: | 2003-06-23 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The role of substrate-binding groups in the mechanism of aspartate-beta-semialdehyde dehydrogenase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1PQP
 
 | | Crystal Structure of the C136S Mutant of Aspartate Semialdehyde Dehydrogenase from Haemophilus influenzae Bound with Aspartate Semialdehyde and Phosphate | | Descriptor: | Aspartate-semialdehyde dehydrogenase, L-HOMOSERINE, PHOSPHATE ION | | Authors: | Blanco, J, Moore, R.A, Faehnle, C.R, Viola, R.E. | | Deposit date: | 2003-06-18 | | Release date: | 2004-08-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The role of substrate-binding groups in the mechanism of aspartate-beta-semialdehyde dehydrogenase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1Q2X
 
 | | Crystal Structure of the E243D Mutant of Aspartate Semialdehyde Dehydrogenase from Haemophilus influenzae bound with substrate aspartate semialdehyde | | Descriptor: | Aspartate-semialdehyde dehydrogenase | | Authors: | Blanco, J, Moore, R.A, Faehnle, C.R, Coe, D.M, Viola, R.E. | | Deposit date: | 2003-07-26 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The role of substrate-binding groups in the mechanism of aspartate-beta-semialdehyde dehydrogenase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
6M2W
 
 | | Structure of RyR1 (Ca2+/Caffeine/ATP/CaM1234/CHL) | | Descriptor: | 5-bromanyl-N-[4-chloranyl-2-methyl-6-(methylcarbamoyl)phenyl]-2-(3-chloranylpyridin-2-yl)pyrazole-3-carboxamide, ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, ... | | Authors: | Ma, R, Haji-Ghassemi, O, Ma, D, Lin, L, Samurkas, A, Van Petegem, F, Yuchi, Z. | | Deposit date: | 2020-03-01 | | Release date: | 2020-09-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for diamide modulation of ryanodine receptor.
Nat.Chem.Biol., 16, 2020
|
|
6LYM
 
 | | Crystal structure of D657A mutant of formylglycinamidine synthetase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Sharma, N, Tanwar, A.S, Anand, R. | | Deposit date: | 2020-02-14 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Mechanism of Coordinated Gating and Signal Transduction in Purine Biosynthetic Enzyme Formylglycinamidine Synthetase.
Acs Catalysis, 12, 2022
|
|
6LYO
 
 | | Crystal Structure of H296A mutant of Formylglycinamidine Synthetase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Sharma, N, Tanwar, A.S, Anand, R. | | Deposit date: | 2020-02-15 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Mechanism of Coordinated Gating and Signal Transduction in Purine Biosynthetic Enzyme Formylglycinamidine Synthetase.
Acs Catalysis, 12, 2022
|
|
6LYK
 
 | | Crystal Structure of R1263A mutant of Formylglycinamidine Synthetase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLUTAMINE, ... | | Authors: | Sharma, N, Tanwar, A.S, Anand, R. | | Deposit date: | 2020-02-14 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of Coordinated Gating and Signal Transduction in Purine Biosynthetic Enzyme Formylglycinamidine Synthetase.
Acs Catalysis, 12, 2022
|
|
1PQU
 
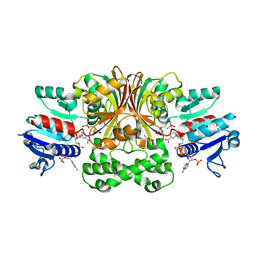 | | Crystal Structure of the H277N Mutant of Aspartate Semialdehyde Dehydrogenase from Haemophilus influenzae Bound with NADP, S-methyl cysteine sulfoxide and cacodylate | | Descriptor: | Aspartate-semialdehyde dehydrogenase, CACODYLATE ION, CYSTEINE, ... | | Authors: | Blanco, J, Moore, R.A, Viola, R.E. | | Deposit date: | 2003-06-19 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The role of substrate-binding groups in the mechanism of aspartate-beta-semialdehyde dehydrogenase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
6LYL
 
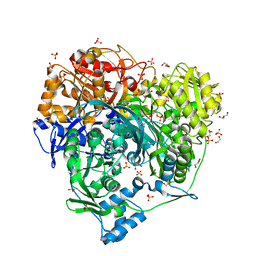 | | Crystal structure of S1052D mutant of Formylglycinamidine synthetase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Sharma, N, Tanwar, A.S, Anand, R. | | Deposit date: | 2020-02-14 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of Coordinated Gating and Signal Transduction in Purine Biosynthetic Enzyme Formylglycinamidine Synthetase.
Acs Catalysis, 12, 2022
|
|
1RBZ
 
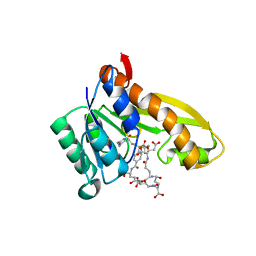 | | Human GAR Tfase complex structure with polyglutamated 10-(trifluoroacetyl)-5,10-dideazaacyclic-5,6,7,8-tetrahydrofolic acid | | Descriptor: | N-{4-4-(2,4-DIAMINO-6-OXO-1,6-DIHYDRO-PYRIMIDIN-5-YL)-1-(2,2,2-TRIFLUORO-1,1-DIHYDROXY-ETHYL)-BUT-2-YL-BENZOYL}-GAMMA-GLUTAMYL-GAMMA-GLUTAMYL-GAMMA-GLUTAMYL-GAMMA-GLUTAMYL-GLUTAMIC ACID, PHOSPHORIBOSYLGLYCINAMIDE FORMYLTRANSFERASE | | Authors: | Zhang, Y, Desharnais, J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2003-11-03 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human GAR Tfase complex structure
To be Published
|
|
1RBQ
 
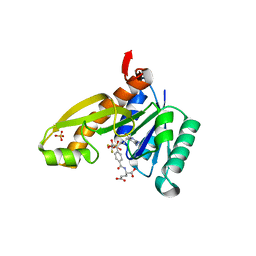 | | Human GAR Tfase complex structure with 10-(trifluoroacetyl)-5,10-dideazaacyclic-5,6,7,8-tetrahydrofolic acid | | Descriptor: | N-{4-[(1R)-4-[(2R,4R,5S)-2,4-DIAMINO-6-OXOHEXAHYDROPYRIMIDIN-5-YL]-1-(2,2,2-TRIFLUORO-1,1-DIHYDROXYETHYL)BUTYL]BENZOYL}-D-GLUTAMIC ACID, PHOSPHATE ION, PHOSPHORIBOSYLGLYCINAMIDE FORMYLTRANSFERASE | | Authors: | Zhang, Y, Desharnais, J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2003-11-03 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Human GAR Tfase complex structure with polyglutamated
10-(trifluoroacetyl)-5,10-dideazaacyclic-5,6,7,8-tetrahydrofolic acid
To be Published
|
|
6JHN
 
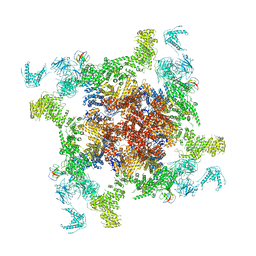 | | Structure of RyR2 (F/C/Ca2+ dataset) | | Descriptor: | CAFFEINE, CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, ... | | Authors: | Chi, X.M, Gong, D.S, Ren, K, Zhou, G.W, Huang, G.X.Y, Lei, J.L, Zhou, Q, Yan, N. | | Deposit date: | 2019-02-18 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Molecular basis for allosteric regulation of the type 2 ryanodine receptor channel gating by key modulators.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6JRR
 
 | | Structure of RyR2 (*F/A/C/L-Ca2+ dataset) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Gong, D.S, Chi, X.M, Zhou, G.W, Huang, G.X.Y, Lei, J.L, Yan, N. | | Deposit date: | 2019-04-05 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Modulation of cardiac ryanodine receptor 2 by calmodulin.
Nature, 572, 2019
|
|
1PS8
 
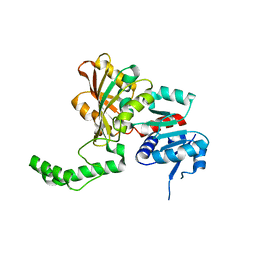 | | Crystal Structure of the R270K Mutant of Aspartate Semialdehyde dehydrogenase from Haemophilus influenzae | | Descriptor: | Aspartate semialdehyde dehydrogenase | | Authors: | Blanco, J, Moore, R.A, Faehnle, C.R, Coe, D.M, Viola, R.E. | | Deposit date: | 2003-06-20 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The role of substrate-binding groups in the mechanism of aspartate-beta-semialdehyde dehydrogenase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
6M4U
 
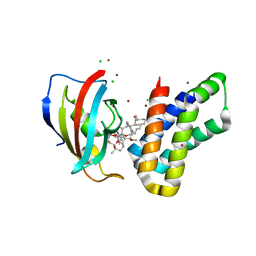 | | Crystal structure of FKBP-FRB T2098L mutant in complex with rapamycin | | Descriptor: | CHLORIDE ION, GLYCEROL, Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Kikuchi, M, Wu, D, Inoue, T, Umehara, T. | | Deposit date: | 2020-03-09 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational design and implementation of a chemically inducible heterotrimerization system.
Nat.Methods, 17, 2020
|
|
6JTA
 
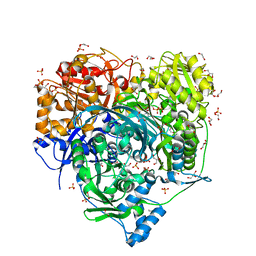 | | Crystal Structure of D464A L465A mutant of FGAM Synthetase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLUTAMINE, ... | | Authors: | Sharma, N, Ahalawat, N, Sandhu, P, Mondal, J, Anand, R. | | Deposit date: | 2019-04-10 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Role of allosteric switches and adaptor domains in long-distance cross-talk and transient tunnel formation.
Sci Adv, 6, 2020
|
|
6MSW
 
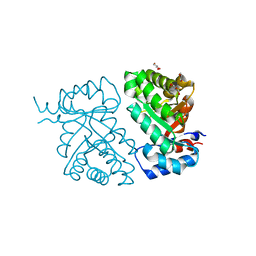 | | Crystal structure of BH1352 2-deoxyribose-5-phosphate from Bacillus halodurans, K184L mutant | | Descriptor: | Deoxyribose-phosphate aldolase, GLYCEROL | | Authors: | Stogios, P.J, Skarina, T, Kim, T, Yim, V, Yakunin, A, Savchenko, A. | | Deposit date: | 2018-10-18 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.169 Å) | | Cite: | Rational engineering of 2-deoxyribose-5-phosphate aldolases for the biosynthesis of (R)-1,3-butanediol.
J.Biol.Chem., 295, 2020
|
|
1PR3
 
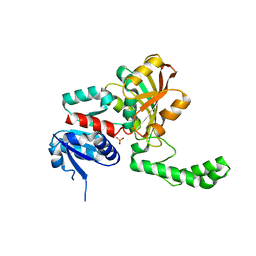 | | Crystal Structure of the R103K Mutant of Aspartate Semialdehyde dehydrogenase from Haemophilus influenzae | | Descriptor: | Aspartate semialdehyde dehydrogenase, PHOSPHATE ION | | Authors: | Blanco, J, Moore, R.A, Faehnle, C.R, Coe, D.M, Viola, R.E. | | Deposit date: | 2003-06-19 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The role of substrate-binding groups in the mechanism of aspartate-beta-semialdehyde dehydrogenase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
6MRC
 
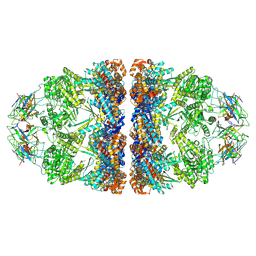 | | ADP-bound human mitochondrial Hsp60-Hsp10 football complex | | Descriptor: | 10 kDa heat shock protein, mitochondrial, 60 kDa heat shock protein, ... | | Authors: | Gomez-Llorente, Y, Jebara, F, Patra, M, Malik, R, Nissemblat, S, Azem, A, Hirsch, J.A, Ubarretxena-Belandia, I. | | Deposit date: | 2018-10-12 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural basis for active single and double ring complexes in human mitochondrial Hsp60-Hsp10 chaperonin.
Nat Commun, 11, 2020
|
|
1S1M
 
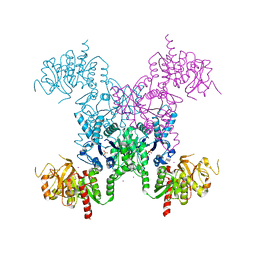 | | Crystal Structure of E. Coli CTP Synthetase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CTP synthase, IODIDE ION, ... | | Authors: | Endrizzi, J.A, Kim, H, Anderson, P.M, Baldwin, E.P. | | Deposit date: | 2004-01-06 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Escherichia coli Cytidine Triphosphate Synthetase, a Nucleotide-Regulated Glutamine Amidotransferase/ATP-Dependent Amidoligase Fusion Protein and Homologue of Anticancer and Antiparasitic Drug Targets
Biochemistry, 43, 2004
|
|
6JT7
 
 | | Crystal structure of 452-453_deletion mutant of FGAM Synthetase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Sharma, N, Ahalawat, N, Sandhu, P, Mondal, J, Anand, R. | | Deposit date: | 2019-04-10 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Role of allosteric switches and adaptor domains in long-distance cross-talk and transient tunnel formation.
Sci Adv, 6, 2020
|
|
1RBY
 
 | | Human GAR Tfase complex structure with 10-(trifluoroacetyl)-5,10-dideazaacyclic-5,6,7,8-tetrahydrofolic acid and substrate beta-GAR | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE, N-{4-[(1R)-4-[(2R,4R,5S)-2,4-DIAMINO-6-OXOHEXAHYDROPYRIMIDIN-5-YL]-1-(2,2,2-TRIFLUORO-1,1-DIHYDROXYETHYL)BUTYL]BENZOYL}-D-GLUTAMIC ACID, PHOSPHORIBOSYLGLYCINAMIDE FORMYLTRANSFERASE | | Authors: | Zhang, Y, Desharnais, J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2003-11-03 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Human GAR Tfase complex structure
To be Published
|
|
1SO5
 
 | | Crystal structure of E112Q mutant of 3-keto-L-gulonate 6-phosphate decarboxylase with bound L-threonohydroxamate 4-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, L-THREONOHYDROXAMATE 4-PHOSPHATE, MAGNESIUM ION | | Authors: | Wise, E.L, Yew, W.S, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-03-12 | | Release date: | 2004-06-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of Enzymatic Activities in the Orotidine 5'-Monophosphate Decarboxylase Suprafamily: Crystallographic Evidence for a Proton Relay System in the Active Site of 3-Keto-l-gulonate 6-Phosphate Decarboxylase(,)
Biochemistry, 43, 2004
|
|
1SO6
 
 | | Crystal structure of E112Q/H136A double mutant of 3-keto-L-gulonate 6-phosphate decarboxylase with bound L-threonohydroxamate 4-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, L-THREONOHYDROXAMATE 4-PHOSPHATE, MAGNESIUM ION | | Authors: | Wise, E.L, Yew, W.S, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-03-12 | | Release date: | 2004-06-08 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Evolution of Enzymatic Activities in the Orotidine 5'-Monophosphate Decarboxylase Suprafamily: Crystallographic Evidence for a Proton Relay System in the Active Site of 3-Keto-l-gulonate 6-Phosphate Decarboxylase(,)
Biochemistry, 43, 2004
|
|
