1F1G
 
 | | Crystal structure of yeast cuznsod exposed to nitric oxide | | Descriptor: | COPPER (II) ION, COPPER-ZINC SUPEROXIDE DISMUTASE, PHOSPHATE ION, ... | | Authors: | Hart, P.J, Ogihara, N.L, Liu, H, Nersissian, A.M, Valentine, J.S, Eisenberg, D. | | Deposit date: | 2000-05-18 | | Release date: | 2002-12-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
3ES8
 
 | | Crystal structure of divergent enolase from Oceanobacillus Iheyensis complexed with Mg and L-malate. | | Descriptor: | (2S)-2-hydroxybutanedioic acid, MAGNESIUM ION, Muconate cycloisomerase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-04 | | Release date: | 2008-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Computation-facilitated assignment of the function in the enolase superfamily: a regiochemically distinct galactarate dehydratase from Oceanobacillus iheyensis .
Biochemistry, 48, 2009
|
|
2VT8
 
 | | Structure of a conserved dimerisation domain within Fbox7 and PI31 | | Descriptor: | PROTEASOME INHIBITOR PI31 SUBUNIT | | Authors: | Kirk, R.J, Murray-Rust, J, Knowles, P.P, Laman, H, McDonald, N.Q. | | Deposit date: | 2008-05-12 | | Release date: | 2008-05-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of a Conserved Dimerization Domain within the F-Box Protein Fbxo7 and the Pi31 Proteasome Inhibitor.
J.Biol.Chem., 283, 2008
|
|
5EIQ
 
 | |
2VQY
 
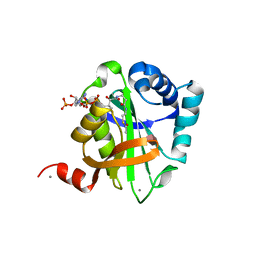 | | Structure of AAC(6')-Ib in complex with Parmomycin and AcetylCoA. | | Descriptor: | AAC(6')-IB, ACETYL COENZYME *A, CALCIUM ION, ... | | Authors: | Vetting, M.W, Park, C.H, Hedge, S.S, Hooper, D.C, Blanchard, J.S. | | Deposit date: | 2008-03-20 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanistic and Structural Analysis of Aminoglycoside N-Acetyltransferase Aac(6')-Ib and its Bifunctional, Fluoroquinolone-Active Aac(6')-Ib-Cr Variant.
Biochemistry, 47, 2008
|
|
2ZR2
 
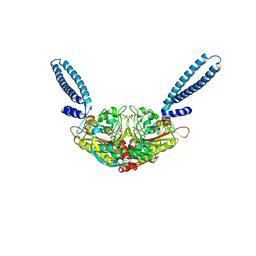 | | Crystal structure of seryl-tRNA synthetase from Pyrococcus horikoshii complexed with ATP | | Descriptor: | ADENOSINE MONOPHOSPHATE, SULFATE ION, Seryl-tRNA synthetase | | Authors: | Itoh, Y, Sekine, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-08-22 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic and mutational studies of seryl-tRNA synthetase from the archaeon Pyrococcus horikoshii.
Rna Biol., 5, 2008
|
|
4M7F
 
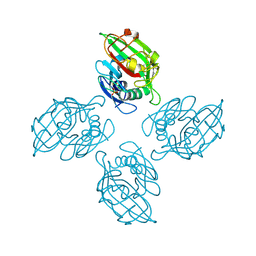 | | Crystal structure of tetrameric fibrinogen-like recognition domain of FIBCD1 with bound ManNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-mannopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Shrive, A.K, Greenhough, T.J, Holmskov, U. | | Deposit date: | 2013-08-12 | | Release date: | 2013-12-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Tetrameric Fibrinogen-like Recognition Domain of Fibrinogen C Domain Containing 1 (FIBCD1) Protein.
J.Biol.Chem., 289, 2014
|
|
4WX2
 
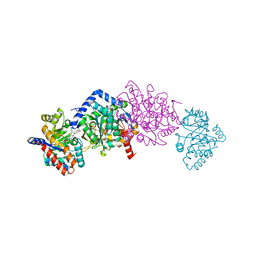 | | Crystal structure of Tryptophan Synthase from Salmonella typhimurium in complex with two F6F molecules in the alpha-site and one F6F molecule in the beta-site | | Descriptor: | 1,2-ETHANEDIOL, 2-{[4-(TRIFLUOROMETHOXY)BENZOYL]AMINO}ETHYL DIHYDROGEN PHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hilario, E, Caulkins, B.G, Young, R.P, Niks, D, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2014-11-13 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Visualizing the tunnel in tryptophan synthase with crystallography: Insights into a selective filter for accommodating indole and rejecting water.
Biochim.Biophys.Acta, 1864, 2016
|
|
8ACN
 
 | | CRYSTAL STRUCTURES OF ACONITASE WITH ISOCITRATE AND NITROISOCITRATE BOUND | | Descriptor: | ACONITASE, IRON/SULFUR CLUSTER, NITROISOCITRIC ACID | | Authors: | Lauble, H, Kennedy, M.C, Beinert, H, Stout, C.D. | | Deposit date: | 1991-05-15 | | Release date: | 1993-10-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of aconitase with isocitrate and nitroisocitrate bound.
Biochemistry, 31, 1992
|
|
2ZV7
 
 | | Lyn Tyrosine Kinase Domain, apo form | | Descriptor: | Tyrosine-protein kinase Lyn | | Authors: | Williams, N.K, Rossjohn, J. | | Deposit date: | 2008-11-04 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of the Lyn Protein Tyrosine Kinase Domain in Its Apo- and Inhibitor-bound State
J.Biol.Chem., 284, 2009
|
|
5ON3
 
 | | Quaternary complex of mutant T252A of E. coli leucyl-tRNA synthetase with tRNA(leu), leucyl-adenylate analogue, and post-transfer editing analogue of leucine in the aminoacylation conformation | | Descriptor: | (2~{S})-~{N}-[(2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-(hydroxymethyl)-4-oxidanyl-oxolan-3-yl]-2-azanyl-4-methyl-pentanamide, 5'-O-(L-leucylsulfamoyl)adenosine, Leucine--tRNA ligase, ... | | Authors: | Palencia, A, Cusack, S. | | Deposit date: | 2017-08-02 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Kinetic Origin of Substrate Specificity in Post-Transfer Editing by Leucyl-tRNA Synthetase.
J. Mol. Biol., 430, 2018
|
|
3TDZ
 
 | | N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex: Structure of a human Cul1WHB-Dcn1P-stapled acetylated Ubc12N complex | | Descriptor: | Cullin-1, DCN1-like protein 1, STAPLED PEPTIDE | | Authors: | Scott, D.C, Monda, J.K, Bennett, E.J, Harper, J.W, Schulman, B.A. | | Deposit date: | 2011-08-11 | | Release date: | 2011-10-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex.
Science, 334, 2011
|
|
5OO9
 
 | | Streptomyces PAC13 (Y55F) with uridine | | Descriptor: | Putative cupin_2 domain-containing isomerase, URIDINE | | Authors: | Chung, C, Michailidou, F. | | Deposit date: | 2017-08-06 | | Release date: | 2017-08-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Pac13 is a Small, Monomeric Dehydratase that Mediates the Formation of the 3'-Deoxy Nucleoside of Pacidamycins.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
6H1B
 
 | | Structure of amide bond synthetase Mcba K483A mutant from Marinactinospora thermotolerans | | Descriptor: | 1-ethanoyl-9~{H}-pyrido[3,4-b]indole-3-carboxylic acid, ADENOSINE MONOPHOSPHATE, Fatty acid CoA ligase | | Authors: | Rowlinson, B, Petchey, M, Cuetos, A, Frese, A, Dannevald, S, Grogan, G. | | Deposit date: | 2018-07-11 | | Release date: | 2018-09-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Broad Aryl Acid Specificity of the Amide Bond Synthetase McbA Suggests Potential for the Biocatalytic Synthesis of Amides.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
4OHO
 
 | | Human GKRP bound to AMG-2668 | | Descriptor: | 5-{[(3S)-3-(prop-1-yn-1-yl)-4-{4-[S-(trifluoromethyl)sulfonimidoyl]phenyl}piperazin-1-yl]sulfonyl}pyridin-2-amine, D-SORBITOL-6-PHOSPHATE, GLYCEROL, ... | | Authors: | Jordan, S.R, Chmait, S. | | Deposit date: | 2014-01-17 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Small molecule disruptors of the glucokinase-glucokinase regulatory protein interaction: 3. Structure-activity relationships within the aryl carbinol region of the N-arylsulfonamido-N'-arylpiperazine series.
J.Med.Chem., 57, 2014
|
|
1W7N
 
 | | Crystal structure of human kynurenine aminotransferase I in PMP form | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, KYNURENINE--OXOGLUTARATE TRANSAMINASE I | | Authors: | Rossi, F, Han, Q, Li, J, Li, J, Rizzi, M. | | Deposit date: | 2004-09-06 | | Release date: | 2004-09-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Human Kynurenine Aminotransferase I
J.Biol.Chem., 279, 2004
|
|
5UUA
 
 | |
1B13
 
 | | CLOSTRIDIUM PASTEURIANUM RUBREDOXIN G10A MUTANT | | Descriptor: | FE (III) ION, PROTEIN (RUBREDOXIN) | | Authors: | Maher, M.J, Guss, J.M, Wilce, M.C.J, Wedd, A.G. | | Deposit date: | 1998-11-26 | | Release date: | 1999-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rubredoxin from Clostridium pasteurianum. Structures of G10A, G43A and G10VG43A mutant proteins. Mutation of conserved glycine 10 to valine causes the 9-10 peptide link to invert.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1B2O
 
 | | CLOSTRIDIUM PASTEURIANUM RUBREDOXIN G10VG43A MUTANT | | Descriptor: | FE (III) ION, PROTEIN (RUBREDOXIN) | | Authors: | Maher, M.J, Guss, J.M, Wilce, M.C.J, Wedd, A.G. | | Deposit date: | 1998-11-30 | | Release date: | 1999-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rubredoxin from Clostridium pasteurianum. Structures of G10A, G43A and G10VG43A mutant proteins. Mutation of conserved glycine 10 to valine causes the 9-10 peptide link to invert.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1B2J
 
 | | CLOSTRIDIUM PASTEURIANUM RUBREDOXIN G43A MUTANT | | Descriptor: | FE (III) ION, PROTEIN (RUBREDOXIN) | | Authors: | Maher, M.J, Guss, J.M, Wilce, M.C.J, Wedd, A.G. | | Deposit date: | 1998-11-27 | | Release date: | 1999-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rubredoxin from Clostridium pasteurianum. Structures of G10A, G43A and G10VG43A mutant proteins. Mutation of conserved glycine 10 to valine causes the 9-10 peptide link to invert.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1W7M
 
 | | Crystal structure of human kynurenine aminotransferase I in complex with L-Phe | | Descriptor: | KYNURENINE--OXOGLUTARATE TRANSAMINASE I, PHENYLALANINE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Rossi, F, Han, Q, Li, J, Li, J, Rizzi, M. | | Deposit date: | 2004-09-06 | | Release date: | 2004-09-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Human Kynurenine Aminotransferase I
J.Biol.Chem., 279, 2004
|
|
8SMX
 
 | | Cryo-EM structure of the human nucleosome core particle in complex with RNF168 and UbcH5c~Ub (UbcH5c chemically conjugated to histone H2A. No density for Ub.) (class 2) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-04-26 | | Release date: | 2024-01-17 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8SMW
 
 | | Cryo-EM structure of the human nucleosome core particle in complex with RNF168 and UbcH5c~Ub (UbcH5c chemically conjugated to histone H2A. No density for Ub.) (class 1) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-04-26 | | Release date: | 2024-01-17 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
7ZTR
 
 | | Crystal structure of the carotenoid-binding protein domain from silkworm Bombyx mori (BmCBP) in the apoform, W232F mutant | | Descriptor: | Carotenoid-binding protein | | Authors: | Sluchanko, N.N, Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Egorkin, N.A, Maksimov, E.G, Popov, V.O. | | Deposit date: | 2022-05-11 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Silkworm carotenoprotein as an efficient carotenoid extractor, solubilizer and transporter.
Int.J.Biol.Macromol., 223, 2022
|
|
8SN6
 
 | | Cryo-EM structure of the human nucleosome core particle in complex with RNF168 and UbcH5c~Ub (UbcH5c chemically conjugated to histone H2A) (class 4) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-04-26 | | Release date: | 2024-01-17 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
