2F14
 
 | | Tne Crystal Structure of the Human Carbonic Anhydrase II in Complex with a Fluorescent Inhibitor | | Descriptor: | 4-(HYDROXYMERCURY)BENZOIC ACID, 5-{[({2-[4-(AMINOSULFONYL)PHENYL]ETHYL}AMINO)CARBONOTHIOYL]AMINO}-2-(6-HYDROXY-3-OXO-3H-XANTHEN-9-YL)BENZOIC ACID, Carbonic anhydrase 2, ... | | Authors: | Alterio, V, Pedone, C, De Simone, G. | | Deposit date: | 2005-11-14 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Carbonic anhydrase inhibitors: X-ray and molecular modeling study for the interaction of a fluorescent antitumor sulfonamide with isozyme II and IX.
J.Am.Chem.Soc., 128, 2006
|
|
2OW9
 
 | | Crystal structure analysis of the MMP13 catalytic domain in complex with specific inhibitor | | Descriptor: | ACETOHYDROXAMIC ACID, BENZYL 6-BENZYL-5,7-DIOXO-6,7-DIHYDRO-5H-[1,3]THIAZOLO[3,2-C]PYRIMIDINE-2-CARBOXYLATE, CALCIUM ION, ... | | Authors: | Pavlovsky, A.G. | | Deposit date: | 2007-02-15 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Discovery and characterization of a novel inhibitor of matrix metalloprotease-13 that reduces cartilage damage in vivo without joint fibroplasia side effects.
J.Biol.Chem., 282, 2007
|
|
2WRZ
 
 | | Crystal structure of an arabinose binding protein with designed serotonin binding site in open, ligand-free state | | Descriptor: | L-ARABINOSE-BINDING PERIPLASMIC PROTEIN | | Authors: | Schreier, B, Stumpp, C, Wiesner, S, Hocker, B. | | Deposit date: | 2009-09-03 | | Release date: | 2009-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Computational Design of Ligand Binding is not a Solved Problem
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7EBC
 
 | | Crystal structure of Isocitrate lyase-1 from Saccaromyces cervisiae | | Descriptor: | Isocitrate lyase, MAGNESIUM ION, TETRAETHYLENE GLYCOL | | Authors: | Hiragi, K, Nishio, K, Moriyama, S, Hamaguchi, T, Mizoguchi, A, Yonekura, K, Tani, K, Mizushima, T. | | Deposit date: | 2021-03-09 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the targeting specificity of ubiquitin ligase for S. cerevisiae isocitrate lyase but not C. albicans isocitrate lyase.
J.Struct.Biol., 213, 2021
|
|
7EBE
 
 | | Crystal structure of Isocitrate lyase-1 from Candida albicans | | Descriptor: | FORMIC ACID, Isocitrate lyase, MAGNESIUM ION | | Authors: | Hiragi, K, Nishio, K, Moriyama, S, Hamaguchi, T, Mizoguchi, A, Yonekura, K, Tani, K, Mizushima, T. | | Deposit date: | 2021-03-09 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural insights into the targeting specificity of ubiquitin ligase for S. cerevisiae isocitrate lyase but not C. albicans isocitrate lyase.
J.Struct.Biol., 213, 2021
|
|
7EBF
 
 | | Cryo-EM structure of Isocitrate lyase-1 from Candida albicans | | Descriptor: | Isocitrate lyase | | Authors: | Hiragi, K, Nishio, K, Moriyama, S, Hamaguchi, T, Mizoguchi, A, Yonekura, K, Tani, K, Mizushima, T. | | Deposit date: | 2021-03-09 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Structural insights into the targeting specificity of ubiquitin ligase for S. cerevisiae isocitrate lyase but not C. albicans isocitrate lyase.
J.Struct.Biol., 213, 2021
|
|
7DXJ
 
 | |
7DXK
 
 | |
2IT8
 
 | |
1Q2B
 
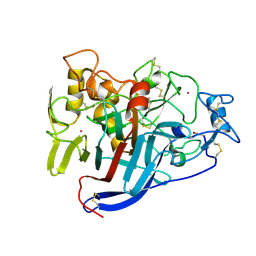 | | CELLOBIOHYDROLASE CEL7A WITH DISULPHIDE BRIDGE ADDED ACROSS EXO-LOOP BY MUTATIONS D241C AND D249C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, EXOCELLOBIOHYDROLASE I | | Authors: | Stahlberg, J, Harris, M, Jones, T.A. | | Deposit date: | 2003-07-24 | | Release date: | 2003-11-25 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Engineering the exo-loop of Trichoderma reesei cellobiohydrolase, Cel7A.
A comparison with Phanerochaete chrysosporium Cel7D.
J.Mol.Biol., 333, 2003
|
|
1Q2E
 
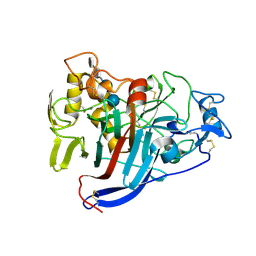 | | CELLOBIOHYDROLASE CEL7A WITH LOOP DELETION 245-252 AND BOUND NON-HYDROLYSABLE CELLOTETRAOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, EXOCELLOBIOHYDROLASE I, ... | | Authors: | Stahlberg, J, Harris, M, Jones, T.A. | | Deposit date: | 2003-07-24 | | Release date: | 2003-11-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Engineering the exo-loop of Trichoderma reesei cellobiohydrolase, Cel7A.
A comparison with Phanerochaete chrysosporium Cel7D
J.Mol.Biol., 333, 2003
|
|
1N1U
 
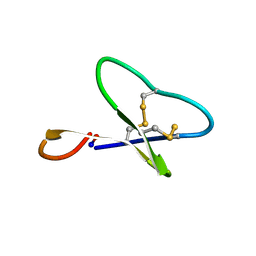 | | NMR structure of [Ala1,15]kalata B1 | | Descriptor: | kalata B1 | | Authors: | Daly, N.L, Clark, R.J, Craik, D.J. | | Deposit date: | 2002-10-20 | | Release date: | 2003-02-25 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Disulfide Folding Pathways of Cystine Knot Proteins. TYING THE KNOT WITHIN THE CIRCULAR BACKBONE OF THE CYCLOTIDES
J.Biol.Chem., 278, 2003
|
|
3DXS
 
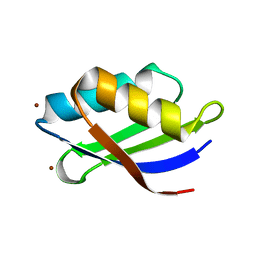 | | Crystal structure of a copper binding domain from HMA7, a P-type ATPase | | Descriptor: | Copper-transporting ATPase RAN1, LITHIUM ION, ZINC ION | | Authors: | Zimmermann, M, Xiao, Z, Clarke, O.B, Gulbis, J.M, Wedd, A.G. | | Deposit date: | 2008-07-25 | | Release date: | 2009-08-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Metal binding affinities of Arabidopsis zinc and copper transporters: selectivities match the relative, but not the absolute, affinities of their amino-terminal domains.
Biochemistry, 48, 2009
|
|
2GKG
 
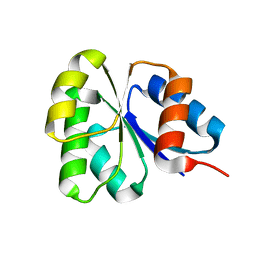 | | Receiver domain from Myxococcus xanthus social motility protein FrzS | | Descriptor: | response regulator homolog | | Authors: | Echols, N, Fraser, J, Merlie, J, Zusman, D, Alber, T. | | Deposit date: | 2006-04-01 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | An atypical receiver domain controls the dynamic polar localization of the Myxococcus xanthus social motility protein FrzS.
Mol.Microbiol., 65, 2007
|
|
2IT7
 
 | |
1RE3
 
 | | Crystal Structure of Fragment D of BbetaD398A Fibrinogen with the Peptide Ligand Gly-His-Arg-Pro-Amide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Fibrinogen alpha/alpha-E chain, ... | | Authors: | Kostelansky, M.S, Betts, L, Gorkun, O.V, Lord, S.T. | | Deposit date: | 2003-11-06 | | Release date: | 2004-05-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | BbetaGlu397 and BbetaAsp398 but not BbetaAsp432 are required for "B:b" interactions.
Biochemistry, 43, 2004
|
|
7EA6
 
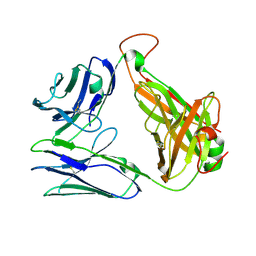 | | Crystal structure of TCR-017 ectodomain | | Descriptor: | T cell receptor 017 alpha chain, T cell receptor 017 beta chain | | Authors: | Nagae, M, Yamasaki, S. | | Deposit date: | 2021-03-06 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.18000245 Å) | | Cite: | Identification of conserved SARS-CoV-2 spike epitopes that expand public cTfh clonotypes in mild COVID-19 patients.
J.Exp.Med., 218, 2021
|
|
1J70
 
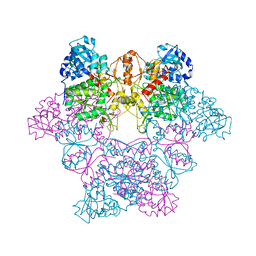 | | CRYSTAL STRUCTURE OF YEAST ATP SULFURYLASE | | Descriptor: | ATP SULPHURYLASE, PHOSPHATE ION, SODIUM ION | | Authors: | Lalor, D.J, Schnyder, T, Saridakis, V, Pilloff, D.E, Dong, A, Tang, H, Leyh, T.S, Pai, E.F. | | Deposit date: | 2001-05-15 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional analysis of a truncated form of Saccharomyces cerevisiae ATP sulfurylase: C-terminal domain essential for oligomer formation but not for activity.
Protein Eng., 16, 2003
|
|
1RE4
 
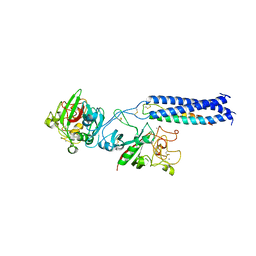 | | Crystal Structure of Fragment D of BbetaD398A Fibrinogen | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Fibrinogen alpha/alpha-E chain, ... | | Authors: | Kostelansky, M.S, Betts, L, Gorkun, O.V, Lord, S.T. | | Deposit date: | 2003-11-06 | | Release date: | 2004-05-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | BbetaGlu397 and BbetaAsp398 but not BbetaAsp432 are required for "B:b" interactions.
Biochemistry, 43, 2004
|
|
2NT4
 
 | | Receiver domain from Myxococcus xanthus social motility protein FrzS (H92F mutant) | | Descriptor: | CHLORIDE ION, Response regulator homolog | | Authors: | Echols, N, Fraser, J, Weisfield, S, Merlie, J, Zusman, D, Alber, T. | | Deposit date: | 2006-11-06 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | An atypical receiver domain controls the dynamic polar localization of the Myxococcus xanthus social motility protein FrzS.
Mol.Microbiol., 65, 2007
|
|
2NT3
 
 | | Receiver domain from Myxococcus xanthus social motility protein FrzS (Y102A Mutant) | | Descriptor: | Response regulator homolog | | Authors: | Fraser, J.S, Echols, N, Merlie, J.P, Zusman, D.R, Alber, T. | | Deposit date: | 2006-11-06 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | An atypical receiver domain controls the dynamic polar localization of the Myxococcus xanthus social motility protein FrzS.
Mol.Microbiol., 65, 2007
|
|
2I6F
 
 | | Receiver domain from Myxococcus xanthus social motility protein FrzS | | Descriptor: | CHLORIDE ION, Response regulator FrzS | | Authors: | Echols, N, Fraser, J, Merlie, J, Zusman, D, Alber, T. | | Deposit date: | 2006-08-28 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An atypical receiver domain controls the dynamic polar localization of the Myxococcus xanthus social motility protein FrzS.
Mol.Microbiol., 65, 2007
|
|
8TMY
 
 | |
8TMZ
 
 | |
3BE7
 
 | | Crystal structure of Zn-dependent arginine carboxypeptidase | | Descriptor: | ARGININE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Meyer, A.J, Freeman, J, Iizuka, M, Bain, K, Rodgers, L, Raushel, F, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-11-16 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional identification of incorrectly annotated prolidases from the amidohydrolase superfamily of enzymes.
Biochemistry, 48, 2009
|
|
