9BDL
 
 | | 80S ribosome with angiogenin | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Korostelev, A.A. | | Deposit date: | 2024-04-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural mechanism of angiogenin activation by the ribosome.
Nature, 630, 2024
|
|
9GBE
 
 | |
9B7F
 
 | | S_SAD structure of HEWL using lossless default compression | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Jakoncic, J, Bernstein, H.J, Soares, A.S, Horvat, K. | | Deposit date: | 2024-03-27 | | Release date: | 2024-04-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Investigation of fast and efficient lossless compression algorithms for macromolecular crystallography experiments.
J.Synchrotron Radiat., 31, 2024
|
|
9F17
 
 | |
7A86
 
 | |
9BCU
 
 | |
9CWL
 
 | |
9F1I
 
 | | Crystal structure of a first-in-class antibody for alpha-1,6-fucosylated prostate-specific antigen, target bound | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(2-azanylethoxy)ethoxy]ethanoic acid, Heavy chain rabbit fab, ... | | Authors: | Halldorsson, S. | | Deposit date: | 2024-04-19 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Development of a first-in-class antibody and a specific assay for alpha-1,6-fucosylated prostate-specific antigen.
Sci Rep, 14, 2024
|
|
9GD2
 
 | | Structure of Chd1 bound to a dinucleosome with a dyad-to-dyad distance of 103 bp. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Resolution of transcription-induced hexasome-nucleosome complexes by Chd1 and FACT.
Mol.Cell, 84, 2024
|
|
9FA8
 
 | | Streptococcal Protein G antibody-binding domain C2 - variant 3 | | Descriptor: | C2 variant 3 | | Authors: | Jonnson, M, Ul Mushtaq, A, Nagy, T.M, von Witting, E, Lofblom, J, Nam, K, Wolf-Watz, M, Hober, S. | | Deposit date: | 2024-05-10 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Cooperative folding as a molecular switch in an evolved antibody binder.
J.Biol.Chem., 300, 2024
|
|
8DKG
 
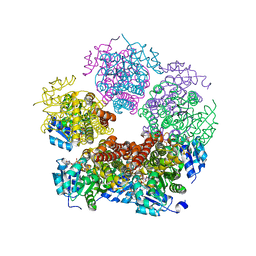 | | Structure of PYCR1 Thr171Met variant complexed with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Isoform 3 of Pyrroline-5-carboxylate reductase 1, mitochondrial, ... | | Authors: | Meeks, K.R, Tanner, J.J. | | Deposit date: | 2022-07-05 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Functional Impact of a Cancer-Related Variant in Human Delta 1 -Pyrroline-5-Carboxylate Reductase 1.
Acs Omega, 8, 2023
|
|
6WX4
 
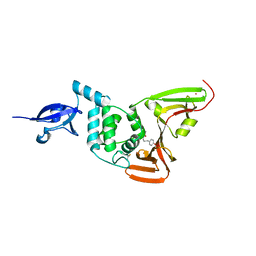 | |
9BLN
 
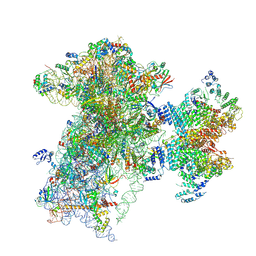 | | The structure of human Pdcd4 bound to the 40S-eIF4A-eIF3-eIF1 complex | | Descriptor: | 40S ribosomal protein S14, ACETIC ACID, Eukaryotic initiation factor 4A-I, ... | | Authors: | Brito Querido, J, Sokabe, M, Diaz-Lopez, I, Gordiyenko, Y, Zuber, P, Yifei, D, Albacete-Albacete, L, Ramakrishnan, V, S Fraser, C. | | Deposit date: | 2024-04-30 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Human tumor suppressor protein Pdcd4 binds at the mRNA entry channel in the 40S small ribosomal subunit.
Nat Commun, 15, 2024
|
|
9G11
 
 | |
7A8G
 
 | |
9B7E
 
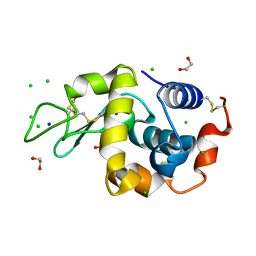 | | S_SAD structure of HEWL using lossy compression data with a compression ratio of 422 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Jakoncic, J, Bernstein, H.J, Soares, A.S, Horvat, K. | | Deposit date: | 2024-03-27 | | Release date: | 2024-04-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Investigation of fast and efficient lossless compression algorithms for macromolecular crystallography experiments.
J.Synchrotron Radiat., 31, 2024
|
|
9BDC
 
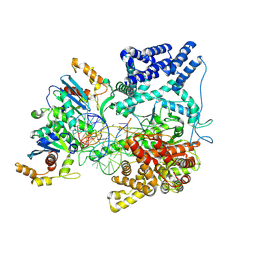 | | Cryo-EM Structure of the TEFM bound Human Mitochondrial Transcription Elongation Complex in a Closed Fingers Domain Conformation | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, Non-Template Strand DNA (NT27mt_+1T), ... | | Authors: | Herbine, K.H, Nayak, A.R, Temiakov, D. | | Deposit date: | 2024-04-11 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Structural basis for substrate binding and selection by human mitochondrial RNA polymerase.
Nat Commun, 15, 2024
|
|
9C6N
 
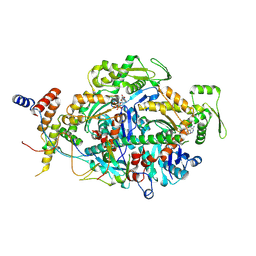 | | ARP module of the human TIP60 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Yang, Z, Mameri, A, Florez Ariza, A.J, Cote, J, Nogales, E. | | Deposit date: | 2024-06-07 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural insights into the human NuA4/TIP60 acetyltransferase and chromatin remodeling complex.
Science, 385, 2024
|
|
9BDW
 
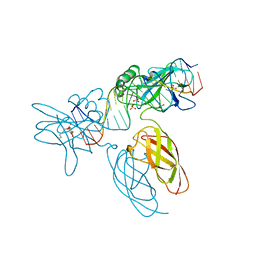 | | NF-kappaB RelA homo-dimer bound to GC-centric kappaB DNA | | Descriptor: | DNA (5'-D(P*AP*CP*TP*GP*GP*GP*AP*AP*GP*TP*TP*CP*CP*AP*GP*TP*GP*AP*T)-3'), DNA (5'-D(P*AP*TP*CP*AP*CP*TP*GP*GP*AP*AP*CP*TP*TP*CP*CP*CP*AP*GP*T)-3'), SULFATE ION, ... | | Authors: | Biswas, T, Shahabi, S, Tsodikov, O.V, Huang, D, Ghosh, G. | | Deposit date: | 2024-04-12 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9EOQ
 
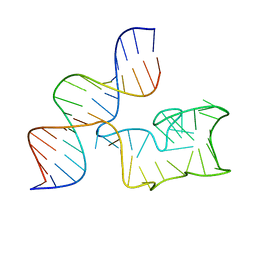 | | Cryo-EM Structure of a 1033 Scaffold Base DNA Origami Nanostructure V4 and TBA | | Descriptor: | DNA (42-MER), DNA (5'-D(P*AP*TP*AP*TP*AP*GP*CP*GP*TP*GP*GP*AP*AP*GP*T)-3') | | Authors: | Ali, K, Georg, K, Volodymyr, M, Johanna, G, Maximilian, N.H, Lukas, K, Simone, C, Hendrik, D. | | Deposit date: | 2024-03-15 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Designing Rigid DNA Origami Templates for Molecular Visualization Using Cryo-EM.
Nano Lett., 24, 2024
|
|
9G4G
 
 | |
9BOQ
 
 | | Human p97/VCP structure with a triazole inhibitor (NSC799462/dodecamer) | | Descriptor: | 3-(2,6-difluoro-4-{[(4P)-5-{[(2S)-hexan-2-yl]sulfanyl}-4-(pyridin-3-yl)-4H-1,2,4-triazol-3-yl]methoxy}phenyl)prop-2-yn-1-yl (1-methylpiperidin-4-yl)carbamate, ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Nandi, P, DeVore, K, Chiu, P.-L. | | Deposit date: | 2024-05-05 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Mechanism of allosteric inhibition of human p97/VCP ATPase and its disease mutant by triazole inhibitors.
Commun Chem, 7, 2024
|
|
9BDU
 
 | | NF-kappaB RelA homo-dimer bound to AT-centric kappaB DNA | | Descriptor: | DNA (5'-D(P*AP*CP*TP*GP*GP*GP*AP*AP*AP*TP*TP*CP*CP*AP*GP*TP*GP*AP*T)-3'), DNA (5'-D(P*AP*TP*CP*AP*CP*TP*GP*GP*AP*AP*TP*TP*TP*CP*CP*CP*AP*GP*T)-3'), SULFATE ION, ... | | Authors: | Biswas, T, Huang, D, Ghosh, G. | | Deposit date: | 2024-04-12 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9CHP
 
 | |
9EOR
 
 | | SARS-CoV2 major protease in complex with a covalent inhibitor SLL12. | | Descriptor: | 3C-like proteinase nsp5, Inhibitor SLL12, POTASSIUM ION | | Authors: | Moche, M, Lennerstrand, J, Nyman, T, Strandback, E, Akaberi, D. | | Deposit date: | 2024-03-15 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Identification of novel and potent inhibitors of SARS-CoV-2 main protease from DNA-encoded chemical libraries.
Antimicrob.Agents Chemother., 68, 2024
|
|
