1KC2
 
 | |
1K51
 
 | | A G55A Mutation Induces 3D Domain Swapping in the B1 Domain of Protein L from Peptostreptococcus magnus | | Descriptor: | Protein L, ZINC ION | | Authors: | O'Neill, J.W, Kim, D.E, Johnsen, K, Baker, D, Zhang, K.Y.J. | | Deposit date: | 2001-10-09 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Single-site mutations induce 3D domain swapping in the B1 domain of protein L from Peptostreptococcus magnus.
Structure, 9, 2001
|
|
1K6S
 
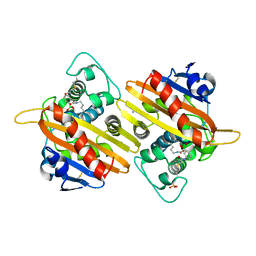 | | STRUCTURE OF THE CLASS D BETA-LACTAMASE OXA-10 IN COMPLEX WITH A PHENYLBORONIC ACID | | Descriptor: | 4-IODO-ACETAMIDO PHENYLBORONIC ACID, Beta-lactamase PSE-2, CALCIUM ION, ... | | Authors: | Kerff, F, Fonze, E, Sauvage, E, Frere, J.M, Charlier, P. | | Deposit date: | 2001-10-17 | | Release date: | 2003-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | CRYSTAL STRUCTURE OF CLASS D BETA-LACTAMASE OXA-10 IN COMPLEX WITH DIFFERENT SUBSTRATES AND ONE INHIBITOR.
To be Published
|
|
1KB6
 
 | |
1KBY
 
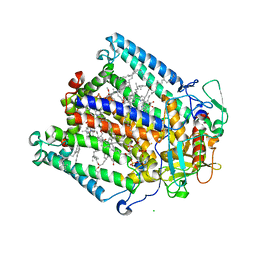 | | Structure of Photosynthetic Reaction Center with bacteriochlorophyll-bacteriopheophytin heterodimer | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Camara-Artigas, A, Magee, C, Goetsch, A, Allen, J.P. | | Deposit date: | 2001-11-07 | | Release date: | 2002-11-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of the heterodimer reaction center from Rhodobacter sphaeroides at 2.55 a resolution.
Photosynth.Res., 74, 2002
|
|
1KAY
 
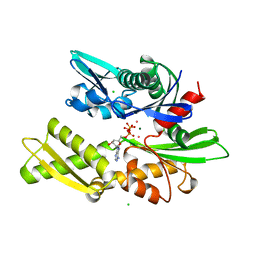 | | 70KD HEAT SHOCK COGNATE PROTEIN ATPASE DOMAIN, K71A MUTANT | | Descriptor: | 70KD HEAT SHOCK COGNATE PROTEIN, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | O'Brien, M.C, Flaherty, K.M, Mckay, D.B. | | Deposit date: | 1996-04-15 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Lysine 71 of the chaperone protein Hsc70 Is essential for ATP hydrolysis.
J.Biol.Chem., 271, 1996
|
|
1K4E
 
 | | CRYSTAL STRUCTURE OF THE CLASS D BETA-LACTAMASES OXA-10 DETERMINED BY MAD PHASING WITH SELENOMETHIONINE | | Descriptor: | Beta-lactamase PSE-2, SULFATE ION | | Authors: | Kerff, F, Fonze, E, Bouillene, F, Frere, J.M, Charlier, P. | | Deposit date: | 2001-10-08 | | Release date: | 2001-10-31 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | STRUCTURE OF CLASS D BETA-LACTAMASE OXA-2
To be Published
|
|
1KBP
 
 | | KIDNEY BEAN PURPLE ACID PHOSPHATASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FE (III) ION, PURPLE ACID PHOSPHATASE, ... | | Authors: | Klabunde, T, Strater, N, Krebs, B. | | Deposit date: | 1995-02-20 | | Release date: | 1996-10-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Mechanism of Fe(III)-Zn(II) purple acid phosphatase based on crystal structures.
J.Mol.Biol., 259, 1996
|
|
1KB0
 
 | |
1KCA
 
 | |
1KMB
 
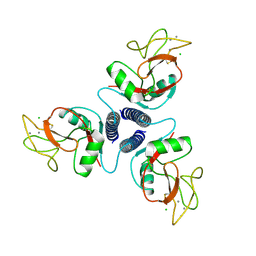 | | SELECTIN-LIKE MUTANT OF MANNOSE-BINDING PROTEIN A | | Descriptor: | CALCIUM ION, CHLORIDE ION, MANNOSE-BINDING PROTEIN-A | | Authors: | Ng, K.K.-S, Weis, W.I. | | Deposit date: | 1996-11-07 | | Release date: | 1997-02-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a selectin-like mutant of mannose-binding protein complexed with sialylated and sulfated Lewis(x) oligosaccharides.
Biochemistry, 36, 1997
|
|
1KQA
 
 | |
1KRQ
 
 | | CRYSTAL STRUCTURE ANALYSIS OF CAMPYLOBACTER JEJUNI FERRITIN | | Descriptor: | ferritin | | Authors: | Hortolan, L, Saintout, N, Granier, G, Langlois d'Estaintot, B, Manigand, C, Mizunoe, Y, Wai, S.N, Gallois, B, Precigoux, G. | | Deposit date: | 2002-01-10 | | Release date: | 2002-02-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | STRUCTURE OF CAMPYLOBACTER JEJUNI FERRITIN AT 2.7 A RESOLUTION
To be Published
|
|
1KCT
 
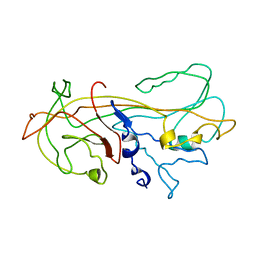 | | ALPHA1-ANTITRYPSIN | | Descriptor: | ALPHA1-ANTITRYPSIN | | Authors: | Song, H.K, Suh, S.W. | | Deposit date: | 1996-08-06 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Crystal structure of an uncleaved alpha 1-antitrypsin reveals the conformation of its inhibitory reactive loop.
FEBS Lett., 377, 1995
|
|
1KUQ
 
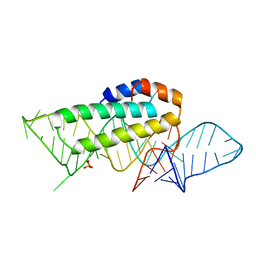 | | CRYSTAL STRUCTURE OF T3C MUTANT S15 RIBOSOMAL PROTEIN IN COMPLEX WITH 16S RRNA | | Descriptor: | 16S RIBOSOMAL RNA FRAGMENT, 30S RIBOSOMAL PROTEIN S15, SULFATE ION | | Authors: | Nikulin, A.D, Tishchenko, S, Revtovich, S, Ehresmann, B, Ehresmann, C, Dumas, P, Garber, M, Nikonov, S, Nevskaya, N. | | Deposit date: | 2002-01-22 | | Release date: | 2003-06-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Role of N-terminal helix in interaction of ribosomal protein S15 with 16S rRNA.
Biochemistry Mosc., 69, 2004
|
|
1KCK
 
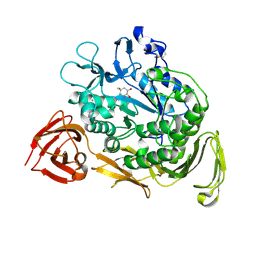 | | Bacillus circulans strain 251 Cyclodextrin glycosyl transferase mutant N193G | | Descriptor: | 1-AMINO-2,3-DIHYDROXY-5-HYDROXYMETHYL CYCLOHEX-5-ENE, CALCIUM ION, CYCLODEXTRIN GLYCOSYLTRANSFERASE, ... | | Authors: | Rozeboom, H.J, Uitdehaag, J.C.M, Dijkstra, B.W. | | Deposit date: | 2001-11-09 | | Release date: | 2002-01-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The remote substrate binding subsite -6 in cyclodextrin-glycosyltransferase controls the transferase activity of the enzyme via an induced-fit mechanism.
J.Biol.Chem., 277, 2002
|
|
1KD3
 
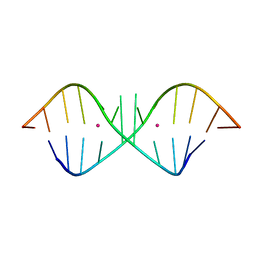 | | The Crystal Structure of r(GGUCACAGCCC)2, Thallium form | | Descriptor: | 5'-R(*GP*GP*UP*CP*AP*CP*AP*GP*CP*CP*C)-3', THALLIUM (I) ION | | Authors: | Kacer, V, Scaringe, S.A, Scarsdale, J.N, Rife, J.P. | | Deposit date: | 2001-11-12 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of r(GGUCACAGCCC)2.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1KOB
 
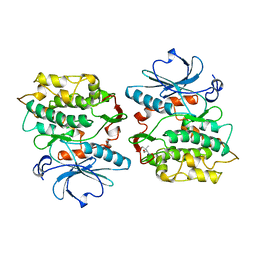 | | TWITCHIN KINASE FRAGMENT (APLYSIA), AUTOREGULATED PROTEIN KINASE DOMAIN | | Descriptor: | TWITCHIN, VALINE | | Authors: | Kobe, B, Heierhorst, J, Feil, S.C, Parker, M.W, Benian, G.M, Weiss, K.R, Kemp, B.E. | | Deposit date: | 1996-06-28 | | Release date: | 1997-03-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Giant protein kinases: domain interactions and structural basis of autoregulation.
EMBO J., 15, 1996
|
|
1KCW
 
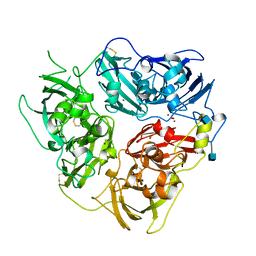 | | X-RAY CRYSTAL STRUCTURE OF HUMAN CERULOPLASMIN AT 3.0 ANGSTROMS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CERULOPLASMIN, COPPER (II) ION, ... | | Authors: | Card, G.L, Zaitsev, V.N, Lindley, P.F. | | Deposit date: | 1996-09-25 | | Release date: | 1997-02-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The X-ray structure of human serum ceruloplasmin at 3.1 angstrom: Nature of the copper centres.
J.Biol.Inorg.Chem., 1, 1996
|
|
1KEI
 
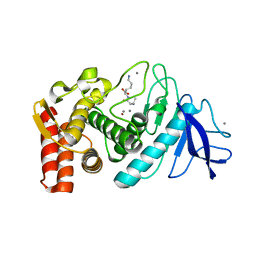 | | Thermolysin (substrate-free) | | Descriptor: | CALCIUM ION, LYSINE, Thermolysin, ... | | Authors: | Senda, M, Senda, T, Kidokoro, S. | | Deposit date: | 2001-11-16 | | Release date: | 2002-11-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure analyses of thermolysin in complex with its inhibitors
To be Published
|
|
1KFO
 
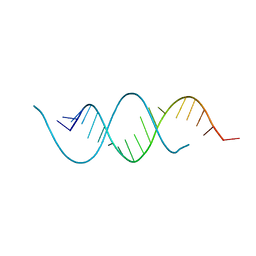 | | CRYSTAL STRUCTURE OF AN RNA HELIX RECOGNIZED BY A ZINC-FINGER PROTEIN: AN 18 BASE PAIR DUPLEX AT 1.6 RESOLUTION | | Descriptor: | 5'-R(*GP*AP*AP*UP*GP*CP*CP*UP*GP*CP*GP*AP*GP*CP*AP*(5BU)P*CP*CP*C)-3' | | Authors: | Lima, S, Hildenbrand, J, Korostelev, A, Hattman, S, Li, H. | | Deposit date: | 2001-11-21 | | Release date: | 2001-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of an RNA helix recognized by a zinc-finger protein: an 18-bp duplex at 1.6 A resolution.
RNA, 8, 2002
|
|
1KEL
 
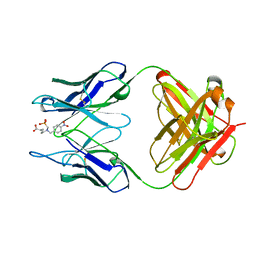 | |
1KEX
 
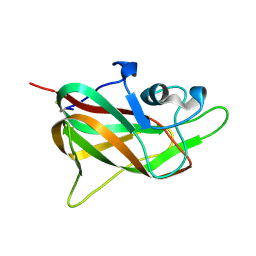 | | Crystal Structure of the b1 Domain of Human Neuropilin-1 | | Descriptor: | Neuropilin-1 | | Authors: | Lee, C.C, Kreusch, A, McMullan, D, Ng, K, Spraggon, G. | | Deposit date: | 2001-11-18 | | Release date: | 2003-01-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Human Neuropilin-1 b1 Domain
Structure, 11, 2003
|
|
1KI3
 
 | | CRYSTAL STRUCTURE OF THYMIDINE KINASE FROM HERPES SIMPLEX VIRUS TYPE I COMPLEXED WITH PENCICLOVIR | | Descriptor: | 9-(4-HYDROXY-3-(HYDROXYMETHYL)BUT-1-YL)GUANINE, SULFATE ION, THYMIDINE KINASE | | Authors: | Champness, J.N, Bennett, M.S, Wien, F, Visse, R, Jarvest, R.L, Summers, W.C, Sanderson, M.R. | | Deposit date: | 1998-05-15 | | Release date: | 1999-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Exploring the active site of herpes simplex virus type-1 thymidine kinase by X-ray crystallography of complexes with aciclovir and other ligands.
Proteins, 32, 1998
|
|
1KI2
 
 | | CRYSTAL STRUCTURE OF THYMIDINE KINASE FROM HERPES SIMPLEX VIRUS TYPE I COMPLEXED WITH GANCICLOVIR | | Descriptor: | 9-(1,3-DIHYDROXY-PROPOXYMETHANE)GUANINE, SULFATE ION, THYMIDINE KINASE | | Authors: | Champness, J.N, Bennett, M.S, Wien, F, Brown, D.G, Visse, R, Sandhu, G, Davies, A, Rizkallah, P.J, Melitz, C, Summers, W.C, Sanderson, M.R. | | Deposit date: | 1998-05-15 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploring the active site of herpes simplex virus type-1 thymidine kinase by X-ray crystallography of complexes with aciclovir and other ligands.
Proteins, 32, 1998
|
|
