2B5S
 
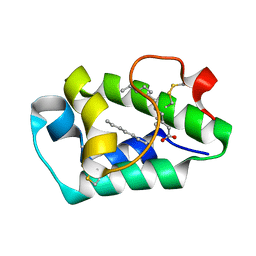 | | Crystal structure of peach Pru p3, the prototypic member of the family of plant non-specific lipid transfer protein pan-allergens | | Descriptor: | HEPTANE, LAURIC ACID, Non-specific lipid transfer protein, ... | | Authors: | Pasquato, N, Berni, R, Folli, C, Folloni, S, Cianci, M, Pantano, S, Helliwell, R.J, Zanotti, G. | | Deposit date: | 2005-09-29 | | Release date: | 2005-11-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Peach Pru p 3, the Prototypic Member of the Family of Plant Non-specific Lipid Transfer Protein Pan-allergens
J.Mol.Biol., 356, 2006
|
|
4MMX
 
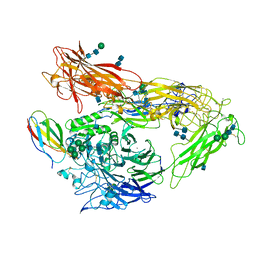 | | Integrin AlphaVBeta3 ectodomain bound to the tenth domain of Fibronectin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fibronectin, ... | | Authors: | van Agthoven, J, Xiong, J, Arnaout, M.A. | | Deposit date: | 2013-09-09 | | Release date: | 2014-03-26 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Structural basis for pure antagonism of integrin alpha V beta 3 by a high-affinity form of fibronectin.
Nat.Struct.Mol.Biol., 21, 2014
|
|
7NZD
 
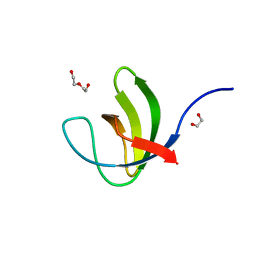 | | Fourth SH3 domain of POSH (Plenty of SH3 Domains protein) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase SH3RF1 | | Authors: | Palencia, A, Bessa, L.M, Jensen, M.R. | | Deposit date: | 2021-03-23 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Visualizing protein breathing motions associated with aromatic ring flipping.
Nature, 602, 2022
|
|
4I9N
 
 | | Crystal structure of rabbit LDHA in complex with AP28161 and AP28122 | | Descriptor: | 6-({2-[(5-chloro-4-{[(2S)-2,3-dihydroxypropyl]oxy}-2-methoxyphenyl)amino]-2-oxoethyl}sulfanyl)pyridine-3-carboxylic acid, 6-[3-(carboxymethoxy)-5-fluorophenyl]pyridine-3-carboxylic acid, L-lactate dehydrogenase A chain | | Authors: | Zhou, T, Kohlmann, A, Stephan, Z.G, Li, F, Commodore, L, Greenfield, M.T, Shakespeare, W.C, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2012-12-05 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Fragment growing and linking lead to novel nanomolar lactate dehydrogenase inhibitors.
J.Med.Chem., 56, 2013
|
|
7NZC
 
 | |
4MQ7
 
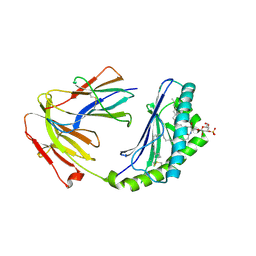 | | Structure of human CD1d-sulfatide | | Descriptor: | (15Z)-N-((1S,2R,3E)-2-HYDROXY-1-{[(3-O-SULFO-BETA-D-GALACTOPYRANOSYL)OXY]METHYL}HEPTADEC-3-ENYL)TETRACOS-15-ENAMIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d, ... | | Authors: | Luoma, A.M, Adams, E.J. | | Deposit date: | 2013-09-15 | | Release date: | 2013-12-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6032 Å) | | Cite: | Crystal Structure of V delta 1 T Cell Receptor in Complex with CD1d-Sulfatide Shows MHC-like Recognition of a Self-Lipid by Human gamma delta T Cells.
Immunity, 39, 2013
|
|
1MYP
 
 | |
3A92
 
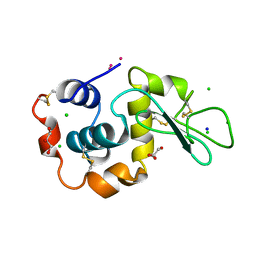 | | Crystal structure of hen egg white lysozyme soaked with 10mM RhCl3 | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C, ... | | Authors: | Abe, S, Koshiyama, T, Ohki, T, Hikage, T, Watanabe, Y, Ueno, T. | | Deposit date: | 2009-10-15 | | Release date: | 2010-03-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Elucidation of Metal-Ion Accumulation Induced by Hydrogen Bonds on Protein Surfaces by Using Porous Lysozyme Crystals Containing Rh(III) Ions as the Model Surfaces
Chemistry, 16, 2010
|
|
4I9U
 
 | | Crystal structure of rabbit LDHA in complex with a fragment inhibitor AP26256 | | Descriptor: | 6-({2-[(5-chloro-2-methoxyphenyl)amino]-2-oxoethyl}sulfanyl)pyridine-3-carboxylic acid, L-lactate dehydrogenase A chain | | Authors: | Zhou, T, Kohlmann, A, Stephan, Z.G, Commodore, L, Greenfield, M.T, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2012-12-05 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment growing and linking lead to novel nanomolar lactate dehydrogenase inhibitors.
J.Med.Chem., 56, 2013
|
|
2XD4
 
 | |
7F96
 
 | | Plasmodium falciparum Prolyl-tRNA Synthetase (PfPRS) in Complex with L-proline and compound L95 | | Descriptor: | PROLINE, Proline--tRNA ligase, ~{N}-[4-[(3~{S})-3-cyano-3-cyclopropyl-2-oxidanylidene-pyrrolidin-1-yl]-6-methyl-pyridin-2-yl]-2-phenyl-ethanamide | | Authors: | Mishra, S, Malhotra, N, Yogavel, M, Sharma, A. | | Deposit date: | 2021-07-04 | | Release date: | 2023-01-04 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.577 Å) | | Cite: | ATP mimetics targeting prolyl-tRNA synthetases as a new avenue for antimalarial drug development
Iscience, 27, 2024
|
|
2DW7
 
 | | Crystal structure of D-tartrate dehydratase from Bradyrhizobium japonicum complexed with Mg++ and meso-tartrate | | Descriptor: | Bll6730 protein, MAGNESIUM ION, S,R MESO-TARTARIC ACID | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2006-08-07 | | Release date: | 2006-12-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evolution of Enzymatic Activities in the Enolase Superfamily: d-Tartrate Dehydratase from Bradyrhizobium japonicum
Biochemistry, 45, 2006
|
|
6XWD
 
 | |
5V97
 
 | |
3L61
 
 | | Crystal structure of substrate-free P450cam at 200 mM [K+] | | Descriptor: | Camphor 5-monooxygenase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lee, Y.-T, Wilson, R.F, Rupniewski, I, Goodin, D.B. | | Deposit date: | 2009-12-22 | | Release date: | 2010-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | P450cam visits an open conformation in the absence of substrate.
Biochemistry, 49, 2010
|
|
5V9F
 
 | |
6MP0
 
 | | Crystal structures of the murine class I major histocompatibility complex H-2Db in complex with the TRP1-M9 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, TRP1-M9 peptide, Beta-2-microglobulin,H-2 class I histocompatibility antigen, ... | | Authors: | Clancy-Thompson, E, Devlin, C.A, Birnbaum, M.E, Dougan, S.K. | | Deposit date: | 2018-10-05 | | Release date: | 2018-11-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Altered Binding of Tumor Antigenic Peptides to MHC Class I Affects CD8+T Cell-Effector Responses.
Cancer Immunol Res, 6, 2018
|
|
4IOL
 
 | | N10-formyltetrahydrofolate synthetase from Moorella thermoacetica with ADP/ZD9 and XPO | | Descriptor: | 2,7-dimethyl-6-[(prop-1-yn-1-ylamino)methyl]quinazolin-4(3H)-one, ADENOSINE-5'-DIPHOSPHATE, Formate--tetrahydrofolate ligase, ... | | Authors: | Stec, B. | | Deposit date: | 2013-01-08 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.563 Å) | | Cite: | Time passes yet errors remain: Comments on the structure of N(10) -formyltetrahydrofolate synthetase.
Protein Sci., 22, 2013
|
|
3L63
 
 | | Crystal structure of camphor-bound P450cam at low [K+] | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Lee, Y.-T, Wilson, R.F, Rupniewski, I, Goodin, D.B. | | Deposit date: | 2009-12-22 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | P450cam visits an open conformation in the absence of substrate.
Biochemistry, 49, 2010
|
|
4IEY
 
 | | Cys-persulfenate bound Cysteine Dioxygenase at pH 7.0 in the presence of Cys, home-source structure | | Descriptor: | Cysteine dioxygenase type 1, FE (II) ION, S-HYDROPEROXYCYSTEINE | | Authors: | Driggers, C.M, Cooley, R.B, Karplus, P.A. | | Deposit date: | 2012-12-13 | | Release date: | 2013-06-26 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Cysteine Dioxygenase Structures from pH4 to 9: Consistent Cys-Persulfenate Formation at Intermediate pH and a Cys-Bound Enzyme at Higher pH.
J.Mol.Biol., 425, 2013
|
|
6FMQ
 
 | | Keap1 - peptide complex | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ACY-SC1-GLU-THR-GLY-GLU-LEU, ... | | Authors: | Talapatra, S.K, Kozielski, F, Wells, G, Georgakopoulos, N.D. | | Deposit date: | 2018-02-02 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Modified Peptide Inhibitors of the Keap1-Nrf2 Protein-Protein Interaction Incorporating Unnatural Amino Acids.
Chembiochem, 19, 2018
|
|
3IHZ
 
 | | Crystal structure of the FK506 binding domain of Plasmodium vivax FKBP35 in complex with FK506 | | Descriptor: | 70 kDa peptidylprolyl isomerase, putative, 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN | | Authors: | Qureshi, I.A, Alag, R, Yoon, H.S, Lescar, J. | | Deposit date: | 2009-07-31 | | Release date: | 2010-06-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | NMR and crystallographic structures of the FK506 binding domain of human malarial parasite Plasmodium vivax FKBP35
Protein Sci., 19, 2010
|
|
3A95
 
 | | Crystal structure of hen egg white lysozyme soaked with 100mM RhCl3 at pH3.8 | | Descriptor: | CHLORIDE ION, Lysozyme C, RHODIUM(III) ION, ... | | Authors: | Abe, S, Koshiyama, T, Ohki, T, Hikage, T, Watanabe, Y, Ueno, T. | | Deposit date: | 2009-10-15 | | Release date: | 2010-03-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Elucidation of Metal-Ion Accumulation Induced by Hydrogen Bonds on Protein Surfaces by Using Porous Lysozyme Crystals Containing Rh(III) Ions as the Model Surfaces
Chemistry, 16, 2010
|
|
6MOM
 
 | | Crystal structure of human Interleukin-1 receptor associated Kinase 4 (IRAK 4, CID 100300) in complex with compound NCC00371481 (BSI 107591) | | Descriptor: | 1,2-ETHANEDIOL, 6-[7-methoxy-6-(1-methyl-1H-pyrazol-4-yl)imidazo[1,2-a]pyridin-3-yl]-N-[(3R)-pyrrolidin-3-yl]pyridin-2-amine, Interleukin-1 receptor-associated kinase 4 | | Authors: | Abendroth, J, Mayclin, S.J, Lorimer, D.D, Starczynowski, D, Hoyt, S, Tawa, G, Thomas, C. | | Deposit date: | 2018-10-04 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Overcoming adaptive therapy resistance in AML by targeting immune response pathways.
Sci Transl Med, 11, 2019
|
|
1R88
 
 | | The crystal structure of Mycobacterium tuberculosis MPT51 (FbpC1) | | Descriptor: | MPT51/MPB51 antigen | | Authors: | Wilson, R.A, Maughan, W.N, Kremer, L, Besra, G.S, Futterer, K. | | Deposit date: | 2003-10-23 | | Release date: | 2003-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The structure of Mycobacterium tuberculosis MPT51 (FbpC1) defines a new family of non-catalytic alpha/beta hydrolases.
J.Mol.Biol., 335, 2004
|
|
