1DIB
 
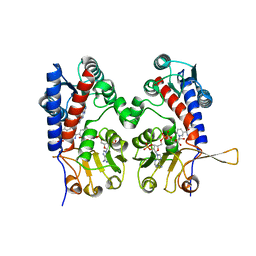 | | HUMAN METHYLENETETRAHYDROFOLATE DEHYDROGENASE / CYCLOHYDROLASE COMPLEXED WITH NADP AND INHIBITOR LY345899 | | Descriptor: | 4-(7-AMINO-9-HYDROXY-1-OXO-3,3A,4,5-TETRAHYDRO-2,5,6,8,9B-PENTAAZA-CYCLOPENTA[A]NAPHTHALEN-2-YL)-PHENYLCARBONYL-GLUTAMI C ACID, METHYLENETETRAHYDROFOLATE DEHYDROGENASE/CYCLOHYDROLASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Schmidt, A, Wu, H, MacKenzie, R.E, Chen, V.J, Bewly, J.R, Ray, J.E, Toth, J.E, Cygler, M. | | Deposit date: | 1999-11-29 | | Release date: | 2000-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of three inhibitor complexes provide insight into the reaction mechanism of the human methylenetetrahydrofolate dehydrogenase/cyclohydrolase.
Biochemistry, 39, 2000
|
|
4H02
 
 | |
2H7R
 
 | |
3BY0
 
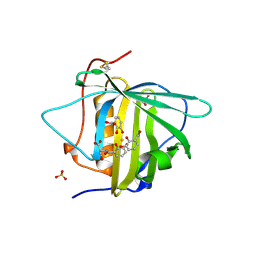 | | Crystal structure of Siderocalin (NGAL, Lipocalin 2) W79A-R81A complexed with Ferric Enterobactin | | Descriptor: | 2,3-DIHYDROXY-BENZOIC ACID, 2-(2,3-DIHYDROXY-BENZOYLAMINO)-3-HYDROXY-PROPIONIC ACID, FE (III) ION, ... | | Authors: | Clifton, M.C, Pizzaro, J.C, Strong, R.K. | | Deposit date: | 2008-01-15 | | Release date: | 2008-09-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.572 Å) | | Cite: | The siderocalin/enterobactin interaction: a link between mammalian immunity and bacterial iron transport.
J.Am.Chem.Soc., 130, 2008
|
|
7EVT
 
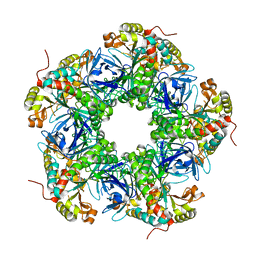 | | Crystal structure of the N-terminal degron-truncated human glutamine synthetase | | Descriptor: | Glutamine synthetase | | Authors: | Chek, M.F, Kim, S.Y, Mori, T, Hakoshima, T. | | Deposit date: | 2021-05-22 | | Release date: | 2021-11-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of N-terminal degron-truncated human glutamine synthetase.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
2ECU
 
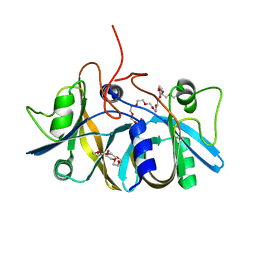 | | Crystal structure of flavin reductase component (HpaC) of 4-hydroxyphenylacetate 3-monooxygenase | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, DODECAETHYLENE GLYCOL, flavin reductase (HpaC) of 4-hydroxyphenylacetate 3-monooxygnease | | Authors: | Kim, S.H, Hisano, T, Iwasaki, W, Ebihara, A, Miki, K. | | Deposit date: | 2007-02-14 | | Release date: | 2008-01-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the flavin reductase component (HpaC) of 4-hydroxyphenylacetate 3-monooxygenase from Thermus thermophilus HB8: Structural basis for the flavin affinity
Proteins, 70, 2008
|
|
1KL8
 
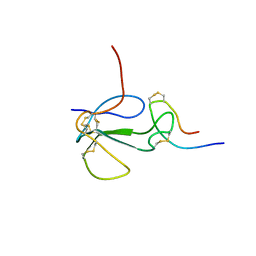 | | NMR STRUCTURAL ANALYSIS OF THE COMPLEX FORMED BETWEEN ALPHA-BUNGAROTOXIN AND THE PRINCIPAL ALPHA-NEUROTOXIN BINDING SEQUENCE ON THE ALPHA7 SUBUNIT OF A NEURONAL NICOTINIC ACETYLCHOLINE RECEPTOR | | Descriptor: | ALPHA-BUNGAROTOXIN, NEURONAL ACETYLCHOLINE RECEPTOR PROTEIN, ALPHA-7 CHAIN | | Authors: | Moise, L, Piserchio, A, Basus, V.J, Hawrot, E. | | Deposit date: | 2001-12-11 | | Release date: | 2002-03-13 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | NMR structural analysis of alpha-bungarotoxin and its complex with the principal alpha-neurotoxin-binding sequence on the alpha 7 subunit of a neuronal nicotinic acetylcholine receptor.
J.Biol.Chem., 277, 2002
|
|
1KC4
 
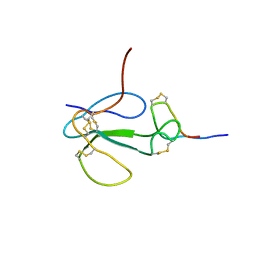 | | NMR Structural Analysis of the Complex Formed Between alpha-Bungarotoxin and the Principal alpha-Neurotoxin Binding Sequence on the alpha7 Subunit of a Neuronal Nicotinic Acetylcholine Receptor | | Descriptor: | alpha-bungarotoxin, neuronal acetylcholine receptor protein, alpha-7 chain | | Authors: | Moise, L, Piserchio, A, Basus, V.J, Hawrot, E. | | Deposit date: | 2001-11-07 | | Release date: | 2002-03-13 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | NMR structural analysis of alpha-bungarotoxin and its complex with the principal alpha-neurotoxin-binding sequence on the alpha 7 subunit of a neuronal nicotinic acetylcholine receptor.
J.Biol.Chem., 277, 2002
|
|
4CPP
 
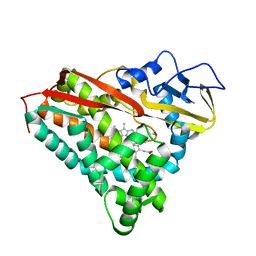 | | CRYSTAL STRUCTURES OF CYTOCHROME P450-CAM COMPLEXED WITH CAMPHANE, THIOCAMPHOR, AND ADAMANTANE: FACTORS CONTROLLING P450 SUBSTRATE HYDROXYLATION | | Descriptor: | ADAMANTANE, CYTOCHROME P450-CAM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Raag, R, Poulos, T.L. | | Deposit date: | 1990-05-18 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structures of cytochrome P-450CAM complexed with camphane, thiocamphor, and adamantane: factors controlling P-450 substrate hydroxylation.
Biochemistry, 30, 1991
|
|
1EP9
 
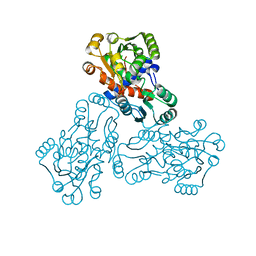 | | HUMAN ORNITHINE TRANSCARBAMYLASE: CRYSTALLOGRAPHIC INSIGHTS INTO SUBSTRATE RECOGNITION AND CONFORMATIONAL CHANGE | | Descriptor: | ORNITHINE TRANSCARBAMYLASE, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER | | Authors: | Shi, D, Morizono, H, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2000-03-28 | | Release date: | 2001-04-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human ornithine transcarbamylase: crystallographic insights into substrate recognition and conformational changes.
Biochem.J., 354, 2001
|
|
2H7S
 
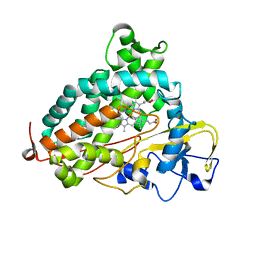 | | L244A mutant of Cytochrome P450cam | | Descriptor: | Cytochrome P450-cam, HEME C | | Authors: | Verras, A, Alian, A, Montellano, P.R. | | Deposit date: | 2006-06-02 | | Release date: | 2006-10-31 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Cytochrome P450 active site plasticity: attenuation of imidazole binding in cytochrome P450cam by an L244A mutation.
Protein Eng.Des.Sel., 19, 2006
|
|
2Z6K
 
 | | Crystal structure of full-length human RPA14/32 heterodimer | | Descriptor: | Replication protein A 14 kDa subunit, Replication protein A 32 kDa subunit | | Authors: | Deng, X, Habel, J.E, Kabaleeswaran, V, Borgstahl, G.E. | | Deposit date: | 2007-08-03 | | Release date: | 2007-12-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Full-length Human RPA14/32 Complex Gives Insights into the Mechanism of DNA Binding and Complex Formation
J.Mol.Biol., 374, 2007
|
|
1OGB
 
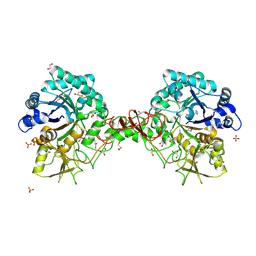 | | Chitinase b from Serratia marcescens mutant D142N | | Descriptor: | CHITINASE B, GLYCEROL, SULFATE ION | | Authors: | Vaaje-Kolstad, G, Houston, D.R, Rao, F.V, Peter, M.G, Synstad, B, van Aalten, D.M.F, Eijsink, V.G.H. | | Deposit date: | 2003-04-29 | | Release date: | 2004-04-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the D142N Mutant of the Family 18 Chitinase Chib from Serratia Marcescens and its Complex with Allosamidin
Biochim.Biophys.Acta, 1696, 2004
|
|
3D26
 
 | | Norwalk P domain A-trisaccharide complex | | Descriptor: | 58 kd capsid protein, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)]beta-D-galactopyranose | | Authors: | Hegde, R, Bu, W. | | Deposit date: | 2008-05-07 | | Release date: | 2008-06-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the receptor binding specificity of Norwalk virus.
J.Virol., 82, 2008
|
|
3D84
 
 | |
1SWJ
 
 | | CORE-STREPTAVIDIN MUTANT W79F AT PH 4.5 | | Descriptor: | CORE-STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Chilkoti, A, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-01-27 | | Release date: | 1999-02-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies of binding site tryptophan mutants in the high-affinity streptavidin-biotin complex.
J.Mol.Biol., 279, 1998
|
|
1SWN
 
 | | CORE-STREPTAVIDIN MUTANT W108F IN COMPLEX WITH BIOTIN AT PH 7.0 | | Descriptor: | BIOTIN, CORE-STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Chilkoti, A, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-01-27 | | Release date: | 1999-02-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural studies of binding site tryptophan mutants in the high-affinity streptavidin-biotin complex.
J.Mol.Biol., 279, 1998
|
|
1SWP
 
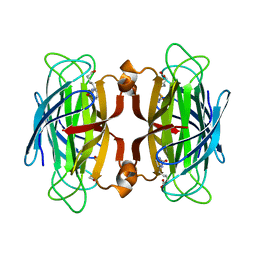 | | CORE-STREPTAVIDIN MUTANT W120F IN COMPLEX WITH BIOTIN AT PH 7.5 | | Descriptor: | BIOTIN, CORE-STREPTAVIDIN, EPI-BIOTIN | | Authors: | Freitag, S, Le Trong, I, Chilkoti, A, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-01-27 | | Release date: | 1999-02-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies of binding site tryptophan mutants in the high-affinity streptavidin-biotin complex.
J.Mol.Biol., 279, 1998
|
|
1SWO
 
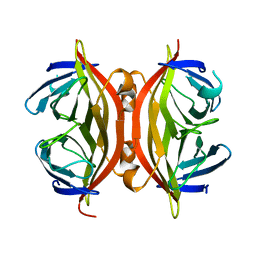 | | CORE-STREPTAVIDIN MUTANT W120F AT PH 7.5 | | Descriptor: | CORE-STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Chilkoti, A, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-01-27 | | Release date: | 1999-02-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural studies of binding site tryptophan mutants in the high-affinity streptavidin-biotin complex.
J.Mol.Biol., 279, 1998
|
|
1SWR
 
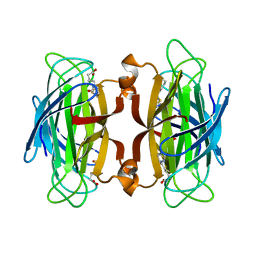 | | CORE-STREPTAVIDIN MUTANT W120A IN COMPLEX WITH BIOTIN AT PH 7.5 | | Descriptor: | BIOTIN, CORE-STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Chilkoti, A, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-01-27 | | Release date: | 1999-02-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural studies of binding site tryptophan mutants in the high-affinity streptavidin-biotin complex.
J.Mol.Biol., 279, 1998
|
|
1U7O
 
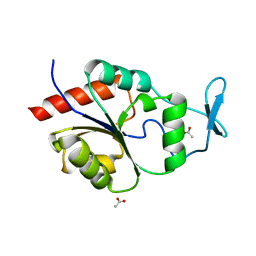 | | Magnesium Dependent Phosphatase 1 (MDP-1) | | Descriptor: | ACETATE ION, magnesium-dependent phosphatase-1 | | Authors: | Peisach, E, Selengut, J.D, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2004-08-04 | | Release date: | 2004-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray crystal structure of the hypothetical phosphotyrosine phosphatase MDP-1 of the haloacid dehalogenase superfamily
Biochemistry, 43, 2004
|
|
2DYU
 
 | |
7KQJ
 
 | | CRYSTAL STRUCTURE OF RAR-RELATED ORPHAN RECEPTOR C (NHIS-RORGT(244-487)-L6-SRC1(678-692) IN COMPLEX WITH A NOVEL TRICYCLIC-CARBOCYLIC RORGT INVERSE AGONIST | | Descriptor: | N-[(3R,3aS,9bS)-9b-[(4-fluorophenyl)sulfonyl]-7-(1,1,1,2,3,3,3-heptafluoropropan-2-yl)-2,3,3a,4,5,9b-hexahydro-1H-cyclopenta[a]naphthalen-3-yl]-2-hydroxy-2-methylpropanamide, Nuclear receptor ROR-gamma | | Authors: | Sack, J.S. | | Deposit date: | 2020-11-16 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.645 Å) | | Cite: | Tricyclic-Carbocyclic ROR gamma t Inverse Agonists-Discovery of BMS-986313.
J.Med.Chem., 64, 2021
|
|
5XBU
 
 | | Crystal structure of GH45 endoglucanase EG27II in apo-form | | Descriptor: | Endo-beta-1,4-glucanase | | Authors: | Nomura, T, Mizutani, K, Iwase, H, Takahashi, N, Mikami, B. | | Deposit date: | 2017-03-21 | | Release date: | 2018-03-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | High-resolution crystal structures of the glycoside hydrolase family 45 endoglucanase EG27II from the snail Ampullaria crossean.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
2A8V
 
 | | RHO TRANSCRIPTION TERMINATION FACTOR/RNA COMPLEX | | Descriptor: | 5'-R(P*CP*CP*C)-3', 5'-R(P*CP*CP*CP*CP*CP*C)-3', RNA BINDING DOMAIN OF RHO TRANSCRIPTION TERMINATION FACTOR | | Authors: | Bogden, C.E, Fass, D, Bergman, N, Nichols, M.D, Berger, J.M. | | Deposit date: | 1998-11-08 | | Release date: | 1999-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structural basis for terminator recognition by the Rho transcription termination factor.
Mol.Cell, 3, 1999
|
|
