7DHL
 
 | | Crystal structure of FGFR3 in complex with pyrimidine derivative | | Descriptor: | 5-[2-(3,5-dimethoxyphenyl)ethyl]-N-[3-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl]pyrimidin-2-amine, Fibroblast growth factor receptor 3 | | Authors: | Echizen, Y, Tateishi, Y, Amano, Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-02-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Synthesis and structure-activity relationships of pyrimidine derivatives as potent and orally active FGFR3 inhibitors with both increased systemic exposure and enhanced in vitro potency.
Bioorg.Med.Chem., 33, 2021
|
|
6FS0
 
 | |
5HW1
 
 | |
3KSU
 
 | | Crystal structure of short-chain dehydrogenase from oenococcus oeni psu-1 | | Descriptor: | 3-oxoacyl-acyl carrier protein reductase | | Authors: | Patskovsky, Y, Toro, R, Gilmore, M, Miller, S, Sauder, J.M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-11-23 | | Release date: | 2009-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Short-Chain Dehydrogenase from Oenococcus Oeni Psu-1
To be Published
|
|
6CSP
 
 | |
7F6F
 
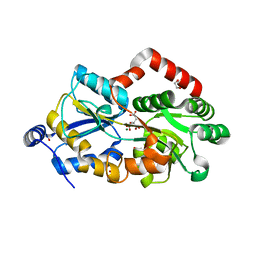 | | Crystal structure of metal-citrate-binding protein (MctA) of ABC transporter endogenously bound to Mg2+-citrate complex (Form II) | | Descriptor: | 1,2-ETHANEDIOL, CARBON DIOXIDE, CITRIC ACID, ... | | Authors: | Kanaujia, S.P, Mandal, S.K, Gogoi, P. | | Deposit date: | 2021-06-25 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and thermodynamic insights into a novel Mg 2+ -citrate-binding protein from the ABC transporter superfamily.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7DIF
 
 | | GH127 beta-L-arabinofuranosidase HypBA1 covalently complexed with beta-L-arabinofuranose-configured cyclophellitol at 1.75-angstrom resolution | | Descriptor: | (1S,2S,3R,4R)-3-(hydroxymethyl)cyclopentane-1,2,4-triol, Non-reducing end beta-L-arabinofuranosidase, POTASSIUM ION, ... | | Authors: | Amaki, S, McGregor, N.G.S, Arakawa, T, Yamada, C, Borlandelli, V, Overkleeft, H.S, Davies, G.J, Fushinobu, S. | | Deposit date: | 2020-11-19 | | Release date: | 2021-01-27 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cysteine Nucleophiles in Glycosidase Catalysis: Application of a Covalent beta-l-Arabinofuranosidase Inhibitor.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
4NXI
 
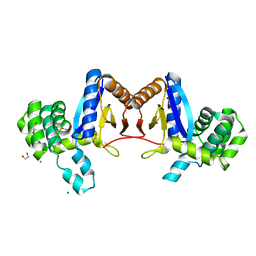 | | Rv2466c Mediates the Activation of TP053 To Kill Replicating and Non-replicating Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Uncharacterized protein | | Authors: | Albesa-Jove, D, Urresti, S, Comino, N, Guerin, M.E. | | Deposit date: | 2013-12-09 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Rv2466c mediates the activation of TP053 to kill replicating and non-replicating Mycobacterium tuberculosis.
Acs Chem.Biol., 9, 2014
|
|
6G3M
 
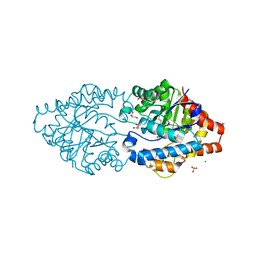 | | Phosphotriesterase PTE_C23M_4 | | Descriptor: | 1-ethyl-1-methyl-cyclohexane, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Dym, O, Aggarwal, N, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Leader, H, Ashani, Y, Goldsmith, M, Greisen, P, Tawfik, D, Sussman, L.J. | | Deposit date: | 2018-03-26 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.665 Å) | | Cite: | Phosphotriesterase
PTE_C23M_4
To Be Published
|
|
5EIH
 
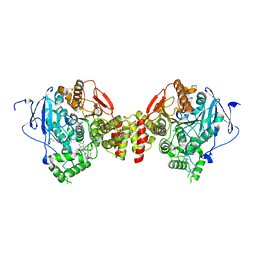 | | mAChE-TZ2/PA5 complex | | Descriptor: | 5-hept-6-ynyl-6-phenyl-phenanthridin-5-ium-3,8-diamine, ACETATE ION, Acetylcholinesterase, ... | | Authors: | Bourne, Y, Marchot, P. | | Deposit date: | 2015-10-29 | | Release date: | 2016-01-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Steric and Dynamic Parameters Influencing In Situ Cycloadditions to Form Triazole Inhibitors with Crystalline Acetylcholinesterase.
J.Am.Chem.Soc., 138, 2016
|
|
7F6O
 
 | |
2IF2
 
 | | Crystal Structure of the Putative Dephospho-CoA Kinase from Aquifex aeolicus, Northeast Structural Genomics Target QR72. | | Descriptor: | 1,2-ETHANEDIOL, Dephospho-CoA kinase, SULFATE ION | | Authors: | Forouhar, F, Hussain, M, Seetharaman, J, Hussain, A, Wu, M, Fang, Y, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M, Rost, B, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-09-19 | | Release date: | 2006-10-03 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: |
|
|
6GBJ
 
 | | Repertoires of functionally diverse enzymes through computational design at epistatic active-site positions | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Parathion hydrolase, ... | | Authors: | Khersonsky, O, Lipsh, R, Avizemer, Z, Goldsmith, M, Ashani, Y, Leader, H, Dym, O, Rogotner, S, Trudeau, D, Tawfik, D.S, Fleishman, S.J. | | Deposit date: | 2018-04-15 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Automated Design of Efficient and Functionally Diverse Enzyme Repertoires.
Mol. Cell, 72, 2018
|
|
1KIZ
 
 | | D100E trichodiene synthase complexed with pyrophosphate | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Rynkiewicz, M.J, Cane, D.E, Christianson, D.W. | | Deposit date: | 2001-12-03 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray crystal structures of D100E trichodiene synthase and its pyrophosphate complex reveal the basis for terpene product diversity.
Biochemistry, 41, 2002
|
|
6GA4
 
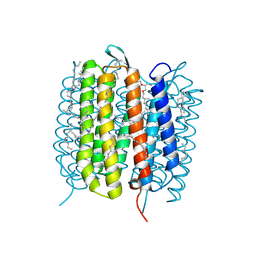 | | Bacteriorhodopsin, 1 ps state, real-space refined against 15% extrapolated map | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
7G16
 
 | | Crystal Structure of human FABP4 in complex with 6-bromo-3-hexyl-2-oxo-3,4-dihydro-1H-quinoline-4-carboxylic acid, i.e. SMILES c12[C@@H]([C@@H](C(=O)Nc1ccc(c2)Br)CCCCCC)C(=O)O with IC50=1.5 microM | | Descriptor: | (3S,4R)-6-bromo-3-hexyl-2-oxo-1,2,3,4-tetrahydroquinoline-4-carboxylic acid, BROMIDE ION, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Hofheinz, W, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
4D6C
 
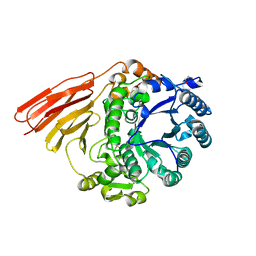 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98)(L19 mutant) | | Descriptor: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
2VLH
 
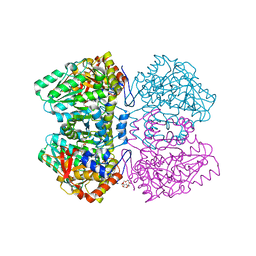 | | Quinonoid intermediate of Citrobacter freundii tyrosine phenol-lyase formed with methionine | | Descriptor: | (2E)-2-{[(Z)-{3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4(1H)-YLIDENE}METHYL]IMINO}-4-(METHYLSULFANYL)BUTANOIC ACID, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, POTASSIUM ION, ... | | Authors: | Milic, D, Demidkina, T.V, Matkovic-Calogovic, D, Antson, A.A. | | Deposit date: | 2008-01-14 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Insights Into the Catalytic Mechanism of Tyrosine Phenol-Lyase from X-Ray Structures of Quinonoid Intermediates.
J.Biol.Chem., 283, 2008
|
|
7FXE
 
 | | Crystal Structure of human FABP4 in complex with 2-(9-benzyl-6-methyl-1,2,3,4-tetrahydrocarbazol-1-yl)acetic acid, i.e. SMILES C12=C(c3c(N1Cc1ccccc1)ccc(c3)C)CCC[C@H]2CC(=O)O with IC50=3.60776 microM | | Descriptor: | Fatty acid-binding protein, adipocyte, SULFATE ION, ... | | Authors: | Ehler, A, Benz, J, Obst, U, ROCHE, N, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
3M7K
 
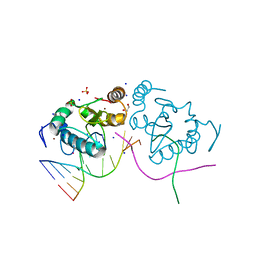 | | Crystal structure of PacI-DNA Enzyme product complex | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*AP*GP*GP*CP*TP*TP*AP*AP*T)-3'), DNA (5'-D(P*TP*AP*AP*GP*CP*CP*TP*C)-3'), ... | | Authors: | Shen, B.W, Stoddard, B.L. | | Deposit date: | 2010-03-16 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Unusual target site disruption by the rare-cutting HNH restriction endonuclease PacI.
Structure, 18, 2010
|
|
4EZ2
 
 | |
3M83
 
 | |
1UB3
 
 | | Crystal Structure of Tetrameric Structure of Aldolase from thermus thermophilus HB8 | | Descriptor: | 1-HYDROXY-PENTANE-3,4-DIOL-5-PHOSPHATE, Aldolase protein | | Authors: | Lokanath, N.K, Miyano, M, Yokoyama, S, Kuramitsu, S, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-03-28 | | Release date: | 2003-04-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of aldolase from Thermus thermophilus HB8 showing the contribution of oligomeric state to thermostability.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
5T2F
 
 | | Structure of the FHA1 domain of Rad53 bound to the BRCT domain of Dbf4 | | Descriptor: | 1,2-ETHANEDIOL, DDK kinase regulatory subunit DBF4,Serine/threonine-protein kinase RAD53 chimeric protein | | Authors: | Guarne, A, Almawi, A.W, Matthews, L.A. | | Deposit date: | 2016-08-23 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | 'AND' logic gates at work: Crystal structure of Rad53 bound to Dbf4 and Cdc7.
Sci Rep, 6, 2016
|
|
7DCF
 
 | | Crystal structure of EHMT2 SET domain in complex with compound 10 | | Descriptor: | 5'-methoxy-6'-(1-methyl-2,3,4,7-tetrahydroazepin-5-yl)spiro[cyclobutane-1,3'-indole]-2'-amine, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYLMETHIONINE, ... | | Authors: | Suzuki, M, Katayama, K. | | Deposit date: | 2020-10-26 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of DS79932728: A Potent, Orally Available G9a/GLP Inhibitor for Treating beta-Thalassemia and Sickle Cell Disease.
Acs Med.Chem.Lett., 12, 2021
|
|
