8R2P
 
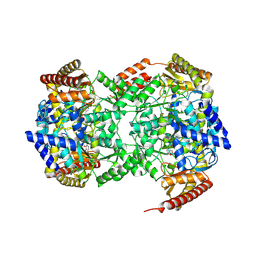 | |
2IJI
 
 | | Structure of F14H mutant of ColE1 Rom protein | | Descriptor: | Regulatory protein rop | | Authors: | Struble, E.B. | | Deposit date: | 2006-09-29 | | Release date: | 2007-10-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New crystal structures of ColE1 Rom and variants resulting from mutation of a surface exposed residue: Implications for RNA-recognition.
Proteins, 72, 2008
|
|
2IMD
 
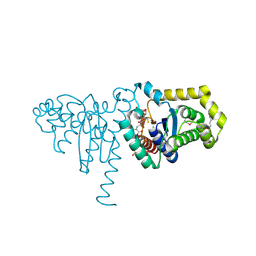 | | Structure of SeMet 2-hydroxychromene-2-carboxylate isomerase (HCCA isomerase) | | Descriptor: | (2S)-2-HYDROXY-2H-CHROMENE-2-CARBOXYLIC ACID, (3E)-4-(2-HYDROXYPHENYL)-2-OXOBUT-3-ENOIC ACID, 2-hydroxychromene-2-carboxylate isomerase, ... | | Authors: | Thompson, L.C, Ladner, J.E, Codreanu, S.G, Harp, J, Gilliland, G.L, Armstrong, R.N. | | Deposit date: | 2006-10-04 | | Release date: | 2007-06-12 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 2-Hydroxychromene-2-carboxylic Acid Isomerase: A Kappa Class Glutathione Transferase from Pseudomonas putida
Biochemistry, 46, 2007
|
|
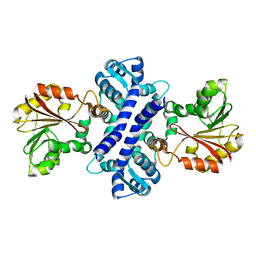
2IP2
 
 | |
2IJH
 
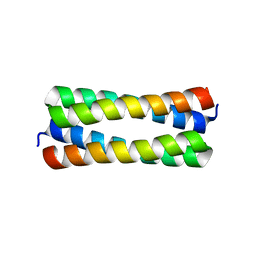 | | Crystal structure analysis of ColE1 ROM mutant F14W | | Descriptor: | Regulatory protein rop | | Authors: | Ladner, J.E. | | Deposit date: | 2006-09-29 | | Release date: | 2007-10-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | New crystal structures of ColE1 Rom and variants resulting from mutation of a surface exposed residue: Implications for RNA-recognition.
Proteins, 72, 2008
|
|
1HK8
 
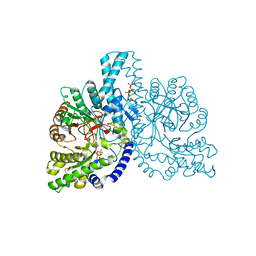 | | STRUCTURAL BASIS FOR ALLOSTERIC SUBSTRATE SPECIFICITY REGULATION IN CLASS III RIBONUCLEOTIDE REDUCTASES: NRDD IN COMPLEX WITH DGTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ANAEROBIC RIBONUCLEOTIDE-TRIPHOSPHATE REDUCTASE, MANGANESE (II) ION, ... | | Authors: | Larsson, K.-M, Andersson, J, Sjoeberg, B.-M, Nordlund, P, Logan, D.T. | | Deposit date: | 2003-03-06 | | Release date: | 2003-03-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A Metal-Binding Site in the Catalytic Subunit of Anaerobic Ribonucleotide Reductase.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1GV9
 
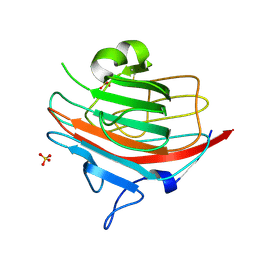 | | p58/ERGIC-53 | | Descriptor: | P58/ERGIC-53, SULFATE ION | | Authors: | Velloso, L.M, Svensson, K, Schneider, G, Pettersson, R.F, Lindqvist, Y. | | Deposit date: | 2002-02-07 | | Release date: | 2002-02-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystal Structure of the Carbohydrate Recognition Domain of P58/Ergic-53, a Protein Involved in Glycoprotein Export from the Endoplasmic Reticulum.
J.Biol.Chem., 277, 2002
|
|
1H7B
 
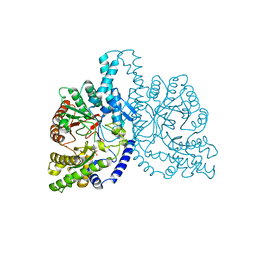 | | Structural basis for allosteric substrate specificity regulation in class III ribonucleotide reductases, native NRDD | | Descriptor: | ANAEROBIC RIBONUCLEOTIDE-TRIPHOSPHATE REDUCTASE LARGE CHAIN, PHOSPHATE ION | | Authors: | Larsson, K.-M, Andersson, J, Sjoeberg, B.-M, Nordlund, P, Logan, D.T. | | Deposit date: | 2001-07-04 | | Release date: | 2002-03-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Basis for Allosteric Substrate Specificty Regulation in Anaerobic Ribonucleotide Reductase
Structure, 9, 2001
|
|
1K39
 
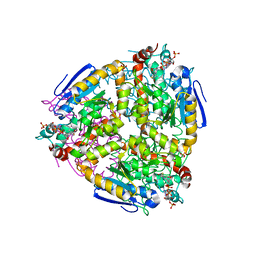 | | The structure of yeast delta3-delta2-enoyl-COA isomerase complexed with octanoyl-COA | | Descriptor: | OCTANOYL-COENZYME A, PHOSPHATE ION, d3,d2-enoyl CoA isomerase ECI1 | | Authors: | Mursula, A.M, Geerlof, A, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 2001-10-02 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: |
|
|
1JAZ
 
 | |
1KO1
 
 | | Crystal structure of gluconate kinase | | Descriptor: | CHLORIDE ION, Gluconate kinase | | Authors: | Kraft, L, Sprenger, G.A, Lindqvist, Y. | | Deposit date: | 2001-12-20 | | Release date: | 2002-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Conformational changes during the catalytic cycle of gluconate kinase as revealed by X-ray crystallography.
J.Mol.Biol., 318, 2002
|
|
1KOF
 
 | | Crystal structure of gluconate kinase | | Descriptor: | Gluconate kinase, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Kraft, L, Sprenger, G.A, Lindqvist, Y. | | Deposit date: | 2001-12-20 | | Release date: | 2002-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational changes during the catalytic cycle of gluconate kinase as revealed by X-ray crystallography.
J.Mol.Biol., 318, 2002
|
|
6QGE
 
 | | Galectin-3C in complex with a pair of enantiomeric ligands: S enantiomer | | Descriptor: | (2~{S},3~{R},4~{S},5~{R},6~{R})-4-[4-(3-fluorophenyl)-1,2,3-triazol-1-yl]-2-[(2~{S})-3-[4-(3-fluorophenyl)-1,2,3-triazol-1-yl]-2-oxidanyl-propyl]sulfanyl-6-(hydroxymethyl)oxane-3,5-diol, Galectin-3 | | Authors: | Manzoni, F, Verteramo, M.L, Oksanen, E, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-01-11 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Interplay between Conformational Entropy and Solvation Entropy in Protein-Ligand Binding.
J. Am. Chem. Soc., 141, 2019
|
|
6QGF
 
 | | Galectin-3C in complex with a pair of enantiomeric ligands: R enantiomer | | Descriptor: | (2~{S},3~{R},4~{S},5~{R},6~{R})-4-[4-(3-fluorophenyl)-1,2,3-triazol-1-yl]-2-[(2~{R})-3-[4-(3-fluorophenyl)-1,2,3-triazol-1-yl]-2-oxidanyl-propyl]sulfanyl-6-(hydroxymethyl)oxane-3,5-diol, Galectin-3 | | Authors: | Manzoni, F, Verteramo, M.L, Oksanen, E, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-01-11 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Interplay between Conformational Entropy and Solvation Entropy in Protein-Ligand Binding.
J. Am. Chem. Soc., 141, 2019
|
|
7R2Y
 
 | | Carbon regulatory PII-like protein SbtB from Synechocystis sp. 6803 in complex with ATP resulting from short ATP soak, conflicting T-loop and C-loop with partial occupancy | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Membrane-associated protein slr1513, SODIUM ION | | Authors: | Selim, K.A, Albrecht, R, Hartmann, M.D. | | Deposit date: | 2022-02-06 | | Release date: | 2023-02-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Carbon signaling protein SbtB possesses atypical redox-regulated apyrase activity to facilitate regulation of bicarbonate transporter SbtA.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6QLN
 
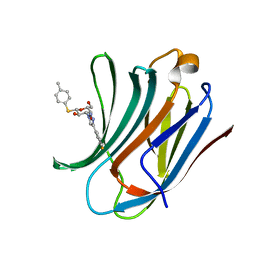 | | Galectin-3C in complex with fluoroaryl triazole monothiogalactoside derivative 2 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{S})-4-[4-(3-fluorophenyl)-1,2,3-triazol-1-yl]-2-(hydroxymethyl)-6-(4-methylphenyl)sulfanyl-oxane-3,5-diol, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-02-01 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure and Energetics of Ligand-Fluorine Interactions with Galectin-3 Backbone and Side-Chain Amides: Insight into Solvation Effects and Multipolar Interactions.
Chemmedchem, 14, 2019
|
|
7R30
 
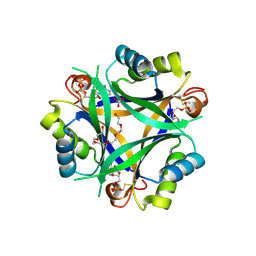 | | Carbon regulatory PII-like protein SbtB from Synechocystis sp. 6803 in complex with ADP and AMP resulting from ADP soak | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BICARBONATE ION, HEXAETHYLENE GLYCOL, ... | | Authors: | Selim, K.A, Albrecht, R, Hartmann, M.D. | | Deposit date: | 2022-02-06 | | Release date: | 2023-02-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Carbon signaling protein SbtB possesses atypical redox-regulated apyrase activity to facilitate regulation of bicarbonate transporter SbtA.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6QLU
 
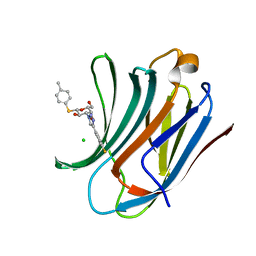 | | Galectin-3C in complex with fluoroaryltriazole monothiogalactoside derivative-8 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{S})-4-[4-(4-fluoranyl-3-methyl-phenyl)-1,2,3-triazol-1-yl]-2-(hydroxymethyl)-6-(4-methylphenyl)sulfanyl-oxane-3,5-diol, CHLORIDE ION, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-02-01 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure and Energetics of Ligand-Fluorine Interactions with Galectin-3 Backbone and Side-Chain Amides: Insight into Solvation Effects and Multipolar Interactions.
Chemmedchem, 14, 2019
|
|
7R2Z
 
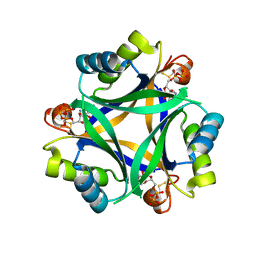 | |
7R32
 
 | | Carbon regulatory PII-like protein SbtB from Synechocystis sp. 6803, delta104 variant, in complex with ADP (co-crystal), tetragonal crystal form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Membrane-associated protein slr1513 | | Authors: | Selim, K.A, Albrecht, R, Hartmann, M.D. | | Deposit date: | 2022-02-06 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Carbon signaling protein SbtB possesses atypical redox-regulated apyrase activity to facilitate regulation of bicarbonate transporter SbtA.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7R31
 
 | | Carbon regulatory PII-like protein SbtB from Synechocystis sp. 6803, C105A+C110A variant, in complex with ATP (co-crystal), tetragonal crystal form | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, Membrane-associated protein slr1513, ... | | Authors: | Selim, K.A, Albrecht, R, Hartmann, M.D. | | Deposit date: | 2022-02-06 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Carbon signaling protein SbtB possesses atypical redox-regulated apyrase activity to facilitate regulation of bicarbonate transporter SbtA.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4EDS
 
 | |
4EDO
 
 | |
6R71
 
 | | Crystal structure of human carbonic anhydrase isozyme XII with 2-(benzenesulfonyl)-4-chloro-N-(2-hydroxyethyl)-5-sulfamoyl-benzamide | | Descriptor: | 1,2-ETHANEDIOL, 4-chloranyl-~{N}-(2-hydroxyethyl)-2-(phenylsulfonyl)-5-sulfamoyl-benzamide, Carbonic anhydrase 12, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2019-03-28 | | Release date: | 2020-04-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Halogenated and di-substituted benzenesulfonamides as selective inhibitors of carbonic anhydrase isoforms.
Eur.J.Med.Chem., 185, 2020
|
|
6R6J
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 2-(benzenesulfonyl)-4-chloro-N-(2-hydroxyethyl)-5-sulfamoyl-benzamide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-chloranyl-~{N}-(2-hydroxyethyl)-2-(phenylsulfonyl)-5-sulfamoyl-benzamide, Carbonic anhydrase 2, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2019-03-27 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Halogenated and di-substituted benzenesulfonamides as selective inhibitors of carbonic anhydrase isoforms.
Eur.J.Med.Chem., 185, 2020
|
|
