6BM9
 
 | |
5G6M
 
 | | Structure of Bacillus subtilis Nitric Oxide Synthase in complex with 7-((3-Aminomethyl)phenoxy)methyl)quinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[[3-(aminomethyl)phenoxy]methyl]quinolin-2-amine, CHLORIDE ION, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2016-06-18 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Targeting Bacterial Nitric Oxide Synthase with Aminoquinoline-Based Inhibitors.
Biochemistry, 55, 2016
|
|
6BN9
 
 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET70 PROTAC | | Descriptor: | Bromodomain-containing protein 4, DNA damage-binding protein 1,DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Nowak, R.P, DeAngelo, S.L, Buckley, D, Bradner, J.E, Fischer, E.S. | | Deposit date: | 2017-11-16 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.382 Å) | | Cite: | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
7M5C
 
 | |
5UAW
 
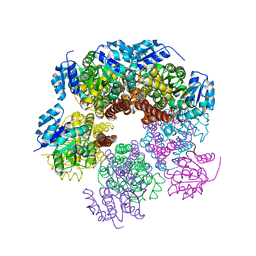 | | Structure of apo human PYCR-1 crystallized in space group P21212 | | Descriptor: | Pyrroline-5-carboxylate reductase 1, mitochondrial, SULFATE ION | | Authors: | Tanner, J.J. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Resolving the cofactor-binding site in the proline biosynthetic enzyme human pyrroline-5-carboxylate reductase 1.
J. Biol. Chem., 292, 2017
|
|
7MQY
 
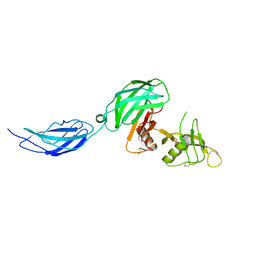 | | Zebrafish CNTN4 APLP2 complex | | Descriptor: | Fusion protein of CNTN4 and APLP2 | | Authors: | Bouyain, S, Karuppan, S.J. | | Deposit date: | 2021-05-07 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Members of the vertebrate contactin and amyloid precursor protein families interact through a conserved interface.
J.Biol.Chem., 298, 2021
|
|
7MRO
 
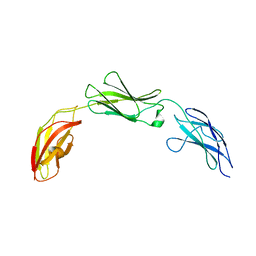 | | Zebrafish CNTN4 FN1-FN3 domains | | Descriptor: | Contactin-4 | | Authors: | Bouyain, S, Karuppan, S.J. | | Deposit date: | 2021-05-07 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Members of the vertebrate contactin and amyloid precursor protein families interact through a conserved interface.
J.Biol.Chem., 298, 2021
|
|
5G1C
 
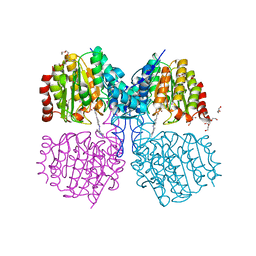 | | Structure of HDAC like protein from Bordetella Alcaligenes bound the photoswitchable pyrazole Inhibitor CEW395 | | Descriptor: | (2E)-N-hydroxy-3-{4-[(E)-(1,3,5-trimethyl-1H-pyrazol-4-yl)diazenyl]phenyl}prop-2-enamide, DI(HYDROXYETHYL)ETHER, HISTONE DEACETYLASE-LIKE AMIDOHYDROLASE, ... | | Authors: | Kraemer, A, Meyer-Almes, F.J, Yildiz, O. | | Deposit date: | 2016-03-24 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Toward Photopharmacological Antimicrobial Chemotherapy Using Photoswitchable Amidohydrolase Inhibitors.
ACS Infect Dis, 3, 2017
|
|
7MRZ
 
 | | Structure of GDF11 bound to fused ActRIIB-ECD and Alk4-ECD with Anti-ActRIIB Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Activin receptor type-2B,Activin receptor type-1B, Fab Heavy Chain, ... | | Authors: | Goebel, E.J, Kattamuri, C, Gipson, G.R, Thompson, T.B. | | Deposit date: | 2021-05-10 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of activin ligand traps using natural sets of type I and type II TGF beta receptors.
Iscience, 25, 2022
|
|
7MRS
 
 | | Zebrafish CNTN4 APPb complex | | Descriptor: | Amyloid-beta A4 protein, Contactin-4 | | Authors: | Bouyain, S, Karuppan, S.J. | | Deposit date: | 2021-05-09 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Members of the vertebrate contactin and amyloid precursor protein families interact through a conserved interface.
J.Biol.Chem., 298, 2021
|
|
7MRM
 
 | | Chicken CNTN3 APP complex | | Descriptor: | Fusion protein of Chicken CNTN3 FN1-FN2 domains and Amyloid-beta A4 protein,Amyloid-beta A4 protein, SULFATE ION | | Authors: | Bouyain, S, Karuppan, S.J. | | Deposit date: | 2021-05-07 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Members of the vertebrate contactin and amyloid precursor protein families interact through a conserved interface.
J.Biol.Chem., 298, 2021
|
|
5G5G
 
 | | Escherichia coli Periplasmic Aldehyde Oxidase | | Descriptor: | ACETATE ION, CHLORIDE ION, DIOXOTHIOMOLYBDENUM(VI) ION, ... | | Authors: | Correia, M.A.S, Otrelo-Cardoso, A.R, Romao, M.J, Santos-Silva, T. | | Deposit date: | 2016-05-25 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Escherichia Coli Periplasmic Aldehyde Oxidoreductase is an Exceptional Member of the Xanthine Oxidase Family of Molybdoenzymes.
Acs Chem.Biol., 11, 2016
|
|
6BL4
 
 | | Crystal Complex of Cyclooxygenase-2 with indomethacin-ethylenediamine-dansyl conjugate | | Descriptor: | 2-[1-(4-chlorobenzene-1-carbonyl)-5-methoxy-2-methyl-1H-indol-3-yl]-N-[2-({[5-(dimethylamino)naphthalen-1-yl]sulfonyl}amino)ethyl]acetamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xu, S, Uddin, M.J, Banerjee, S, Marnett, L.J. | | Deposit date: | 2017-11-09 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Fluorescent indomethacin-dansyl conjugates utilize the membrane-binding domain of cyclooxygenase-2 to block the opening to the active site.
J.Biol.Chem., 294, 2019
|
|
5G31
 
 | |
7MRK
 
 | | Chicken CNTN4 APP complex | | Descriptor: | Amyloid-beta A4 protein, Contactin-4, DI(HYDROXYETHYL)ETHER | | Authors: | Bouyain, S, Karuppan, S.J. | | Deposit date: | 2021-05-07 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Members of the vertebrate contactin and amyloid precursor protein families interact through a conserved interface.
J.Biol.Chem., 298, 2021
|
|
7MWT
 
 | |
7MRN
 
 | | Mouse CNTN5 APP complex | | Descriptor: | Contactin-5, N-APP | | Authors: | Bouyain, S, Karuppan, S.J. | | Deposit date: | 2021-05-07 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Members of the vertebrate contactin and amyloid precursor protein families interact through a conserved interface.
J.Biol.Chem., 298, 2021
|
|
5UCW
 
 | |
6BNB
 
 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET57 PROTAC | | Descriptor: | Bromodomain-containing protein 4, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Nowak, R.P, DeAngelo, S.L, Buckley, D, Ishoey, M, He, Z, Zhang, T, Bradner, J.E, Fischer, E.S. | | Deposit date: | 2017-11-16 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (6.343 Å) | | Cite: | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
6HCV
 
 | | Crystal Structure of Lysyl-tRNA Synthetase from Plasmodium falciparum complexed with a chromone ligand | | Descriptor: | 6-fluoranyl-~{N}-[(1-oxidanylcyclohexyl)methyl]-4-oxidanylidene-chromene-2-carboxamide, Lysine--tRNA ligase | | Authors: | Robinson, D.A, Baragana, B, Norcross, N, Forte, B, Walpole, C, Gilbert, I.H. | | Deposit date: | 2018-08-16 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Lysyl-tRNA synthetase as a drug target in malaria and cryptosporidiosis.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6HB5
 
 | | Crystal structure of E. coli tyrRS in complex with 5'-O-(N-L-tyrosyl)sulfamoyl-cytidine | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Tyrosine--tRNA ligase, ... | | Authors: | De Graef, S, Pang, L, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2018-08-09 | | Release date: | 2019-04-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Comparative analysis of pyrimidine substituted aminoacyl-sulfamoyl nucleosides as potential inhibitors targeting class I aminoacyl-tRNA synthetases.
Eur.J.Med.Chem., 173, 2019
|
|
5UGH
 
 | | Crystal structure of Mat2a bound to the allosteric inhibitor PF-02929366 | | Descriptor: | 2-(7-chloro-5-phenyl[1,2,4]triazolo[4,3-a]quinolin-1-yl)-N,N-dimethylethan-1-amine, S-adenosylmethionine synthase isoform type-2 | | Authors: | Kaiser, S.E, Feng, J, Stewart, A.E. | | Deposit date: | 2017-01-08 | | Release date: | 2017-05-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.062 Å) | | Cite: | Targeting S-adenosylmethionine biosynthesis with a novel allosteric inhibitor of Mat2A.
Nat. Chem. Biol., 13, 2017
|
|
5TY7
 
 | | Crystal structure of wild-type S. aureus penicillin binding protein 4 (PBP4) in complex with nafcillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2-ethoxynaphthalen-1-yl)carbonyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Penicillin-binding protein 4, SODIUM ION, ... | | Authors: | Alexander, J.A.N, Strynadka, N.C.J. | | Deposit date: | 2016-11-18 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.894 Å) | | Cite: | Structural and kinetic analysis of penicillin-binding protein 4 (PBP4)-mediated antibiotic resistance inStaphylococcus aureus.
J. Biol. Chem., 2018
|
|
7MLP
 
 | | Crystal structure of ricin A chain in complex with 5-(2,6-dimethylphenyl)thiophene-2-carboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, 5-(2,6-dimethylphenyl)thiophene-2-carboxylic acid, Ricin, ... | | Authors: | Harijan, R.K, Li, X.P, Cao, B, Augeri, D, Bonanno, J.B, Almo, S.C, Tumer, N.E, Schramm, V.L. | | Deposit date: | 2021-04-28 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Synthesis and Structural Characterization of Ricin Inhibitors Targeting Ribosome Binding Using Fragment-Based Methods and Structure-Based Design.
J.Med.Chem., 64, 2021
|
|
5UHK
 
 | |
