6GD8
 
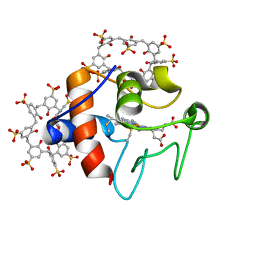 | | Cytochrome c in complex with Sulfonato-calix[8]arene, P31 form | | Descriptor: | Cytochrome c iso-1, HEME C, sulfonato-calix[8]arene | | Authors: | Rennie, M.L, Fox, G.C, Crowley, P.B. | | Deposit date: | 2018-04-23 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Auto-regulated Protein Assembly on a Supramolecular Scaffold.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
7QAT
 
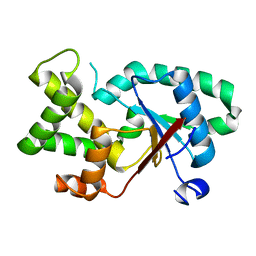 | |
5ZT1
 
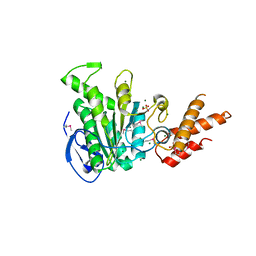 | | Structure of the bacterial pathogens ATPase with substrate ATP gamma S | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Probable ATP synthase SpaL/MxiB, ... | | Authors: | Gao, X.P, Mu, Z.X, Cui, S. | | Deposit date: | 2018-05-01 | | Release date: | 2018-05-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.114 Å) | | Cite: | Structural Insight Into Conformational Changes Induced by ATP Binding in a Type III Secretion-Associated ATPase FromShigella flexneri
Front Microbiol, 9, 2018
|
|
7QAW
 
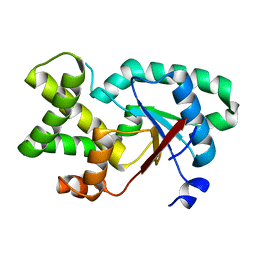 | |
7QAX
 
 | |
7QAU
 
 | |
7QAQ
 
 | |
7QB8
 
 | |
7QAY
 
 | |
6FQL
 
 | | Crystal structure of Danio rerio Lin41 filamin-NHL domains in complex with mab-10 3'UTR 13mer RNA | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase TRIM71, RNA (5'-R(*UP*GP*CP*AP*UP*UP*UP*AP*AP*UP*GP*CP*A)-3') | | Authors: | Kumari, P, Aeschimann, F, Gaidatzis, D, Keusch, J.J, Ghosh, P, Neagu, A, Pachulska-Wieczorek, K, Bujnicki, J.M, Gut, H, Grosshans, H, Ciosk, R. | | Deposit date: | 2018-02-14 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Evolutionary plasticity of the NHL domain underlies distinct solutions to RNA recognition.
Nat Commun, 9, 2018
|
|
4DHI
 
 | | Structure of C. elegans OTUB1 bound to human UBC13 | | Descriptor: | Ubiquitin thioesterase otubain-like, Ubiquitin-conjugating enzyme E2 N | | Authors: | Wiener, R, Zhang, X, Wang, T, Wolberger, C. | | Deposit date: | 2012-01-27 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The mechanism of OTUB1-mediated inhibition of ubiquitination.
Nature, 483, 2012
|
|
3BC5
 
 | | X-ray crystal structure of human ppar gamma with 2-(5-(3-(2-(5-methyl-2-phenyloxazol-4-yl)ethoxy)benzyl)-2-phenyl-2h-1,2,3-triazol-4-yl)acetic acid | | Descriptor: | (5-{3-[2-(5-methyl-2-phenyl-1,3-oxazol-4-yl)ethoxy]benzyl}-2-phenyl-2H-1,2,3-triazol-4-yl)acetic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Muckelbauer, J.K. | | Deposit date: | 2007-11-12 | | Release date: | 2008-11-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Design, synthesis and structure-activity relationships of azole acids as novel, potent dual PPAR alpha/gamma agonists.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6FTP
 
 | |
9DRX
 
 | | Human GABAA receptor of beta2-alpha1-beta2-alpha1-gamma2 subtype in complex with GABA plus Lamotrigine | | Descriptor: | (6M)-6-(2,3-dichlorophenyl)-1,2,4-triazine-3,5-diamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, J, Hibbs, R.E, Noviello, C.M. | | Deposit date: | 2024-09-26 | | Release date: | 2025-01-22 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Resolving native GABA A receptor structures from the human brain.
Nature, 638, 2025
|
|
4NK2
 
 | | Crystal structure of Hell's gate globin IV | | Descriptor: | Hemoglobin-like protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jamil, F. | | Deposit date: | 2013-11-12 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of truncated haemoglobin from an extremely thermophilic and acidophilic bacterium.
J.Biochem., 156, 2014
|
|
5AG6
 
 | | CRYSTAL STRUCTURE OF LEISHMANIA MAJOR N-MYRISTOYLTRANSFERASE (NMT) WITH BOUND MYRISTOYL-COA AND A THIAZOLIDINONE LIGAND | | Descriptor: | (2R)-2-(4-hydroxy-3-methoxyphenyl)-3-(pyridin-2-ylmethyl)-1,3-thiazolidin-4-one, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA | | Authors: | Robinson, D.A, Spinks, D, Smith, V.C, Thompson, S, Smith, A, Torrie, L.S, McElroy, S.P, Brand, S, Brenk, R, Frearson, J.A, Read, K.D, Wyatt, P.G, Gilbert, I.H. | | Deposit date: | 2015-01-29 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of Small-Molecule Trypanosoma Brucei N-Myristoyltransferase Inhibitors: Discovery and Optimisation of a Novel Binding Mode.
Chemmedchem, 10, 2015
|
|
6UWZ
 
 | | Cryo-EM structure of Torpedo acetylcholine receptor in complex with alpha-bungarotoxin | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine receptor subunit alpha, ... | | Authors: | Rahman, M.M, Teng, J, Worrell, B.T, Noveillo, C.M, Lee, M, Karlin, A, Stowell, M, Hibbs, R.E. | | Deposit date: | 2019-11-06 | | Release date: | 2020-04-15 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structure of the Native Muscle-type Nicotinic Receptor and Inhibition by Snake Venom Toxins.
Neuron, 106, 2020
|
|
1GX2
 
 | |
4DHJ
 
 | | The structure of a ceOTUB1 ubiquitin aldehyde UBC13~Ub complex | | Descriptor: | Ubiquitin, Ubiquitin aldehyde, Ubiquitin thioesterase otubain-like, ... | | Authors: | Wiener, R, Zhang, X, Wang, T, Wolberger, C. | | Deposit date: | 2012-01-27 | | Release date: | 2012-02-22 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The mechanism of OTUB1-mediated inhibition of ubiquitination.
Nature, 483, 2012
|
|
5VO0
 
 | | Structure of a TRAF6-Ubc13~Ub complex | | Descriptor: | POTASSIUM ION, TNF receptor-associated factor 6, Ubiquitin, ... | | Authors: | Middleton, A.J, Day, C.L. | | Deposit date: | 2017-05-01 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | The activity of TRAF RING homo- and heterodimers is regulated by zinc finger 1.
Nat Commun, 8, 2017
|
|
6O72
 
 | | Structure of the TRPM8 cold receptor by single particle electron cryo-microscopy, TC-I 2014-bound state | | Descriptor: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(heptanoyloxy)methyl]ethyl octadecanoate, 3-{7-(trifluoromethyl)-5-[2-(trifluoromethyl)phenyl]-1H-benzimidazol-2-yl}-1-oxa-2-azaspiro[4.5]dec-2-ene, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Diver, M.M, Cheng, Y, Julius, D. | | Deposit date: | 2019-03-07 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into TRPM8 inhibition and desensitization.
Science, 365, 2019
|
|
1E2J
 
 | | The nucleoside binding site of Herpes simplex type 1 thymidine kinase analyzed by X-ray crystallography | | Descriptor: | SULFATE ION, THYMIDINE, THYMIDINE KINASE | | Authors: | Vogt, J, Scapozza, L, Schulz, G.E. | | Deposit date: | 2000-05-23 | | Release date: | 2000-11-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nucleoside Binding Site of Herpes Simplex Type 1 Thymidine Kinase Analyzed by X-Ray Crystallography
Proteins: Struct.,Funct., Genet., 41, 2000
|
|
3IF1
 
 | | Crystal structure of 237mAb in complex with a GalNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, Immunoglobulin heavy chain (IgG2a), Immunoglobulin light chain (IgG2a), ... | | Authors: | Brooks, C.L, Evans, S.V, Borisova, S.N. | | Deposit date: | 2009-07-23 | | Release date: | 2010-06-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Antibody recognition of a unique tumor-specific glycopeptide antigen.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2UW6
 
 | | Structure of PKA-PKB chimera complexed with (S)-2-(4-chloro-phenyl)- 2-(4-1H-pyrazol-4-yl)-phenyl)-ethylamine | | Descriptor: | (2S)-2-(4-CHLOROPHENYL)-2-[4-(1H-PYRAZOL-4-YL)PHENYL]ETHANAMINE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, CAMP-DEPENDENT PROTEIN KINASE, ... | | Authors: | Davies, T.G, Saxty, G, Woodhead, S.J, Berdini, V, Verdonk, M.L, Wyatt, P.G, Boyle, R.G, Barford, D, Downham, R, Garrett, M.D, Carr, R.A. | | Deposit date: | 2007-03-19 | | Release date: | 2007-05-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Identification of Inhibitors of Protein Kinase B Using Fragment-Based Lead Discovery
J.Med.Chem., 50, 2007
|
|
1M8S
 
 | | Crystal Structures of Cadmium-binding Acidic Phospholipase A2 from the Venom of Agkistrodon halys pallas at 1.9 Resolution (crystal grown at pH 5.9) | | Descriptor: | 1,4-BUTANEDIOL, CADMIUM ION, phospholipase a2 | | Authors: | Xu, S, Gu, L, Zhou, Y, Lin, Z. | | Deposit date: | 2002-07-25 | | Release date: | 2003-02-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of cadmium-binding acidic phospholipase A(2) from the venom of Agkistrodon halys Pallas at 1.9A resolutio
Biochem.Biophys.Res.Commun., 300, 2003
|
|
