3U3R
 
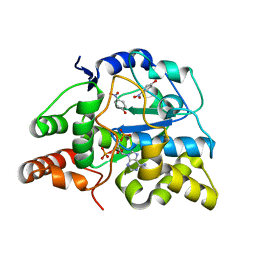 | | Crystal structure of D249G mutated Human SULT1A1 bound to PAP and P-NITROPHENOL | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, P-NITROPHENOL, Sulfotransferase 1A1 | | Authors: | Guttman, C, Berger, I, Aharoni, A, Zarivach, R. | | Deposit date: | 2011-10-06 | | Release date: | 2011-11-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | The molecular basis for the broad substrate specificity of human sulfotransferase 1A1.
Plos One, 6, 2011
|
|
3U45
 
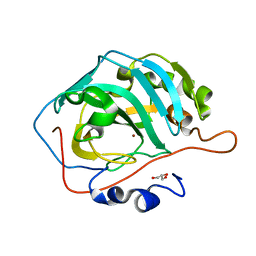 | | Human Carbonic Anhydrase II V143A | | Descriptor: | Carbonic anhydrase 2, GLYCEROL, ZINC ION | | Authors: | West, D, Mckenna, R. | | Deposit date: | 2011-10-07 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Structural Modification of the hydrophobic pocket in Human Carbonic Anhydrase II
To be Published
|
|
3HD1
 
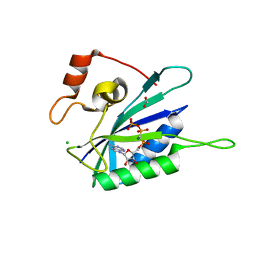 | | Crystal structure of E. coli HPPK(N10A) in complex with MgAMPCPP | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Blaszczyk, J, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2009-05-06 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Role of loop coupling in enzymatic catalysis and conformational dynamics
To be Published
|
|
3U2H
 
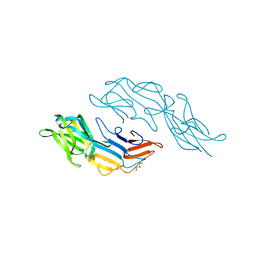 | | Crystal structure of the C-terminal DUF1608 domain of the Methanosarcina acetivorans S-layer (MA0829) protein | | Descriptor: | GLYCEROL, S-layer protein MA0829 | | Authors: | Chan, S, Phan, T, Ahn, C.J, Shin, A, Rohlin, L, Gunsalus, R.P, Arbing, M.A. | | Deposit date: | 2011-10-03 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structure of the surface layer of the methanogenic archaean Methanosarcina acetivorans.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3GVR
 
 | |
7LRS
 
 | |
3U5L
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a benzo-triazepine ligand (BzT-7) | | Descriptor: | 1,2-ETHANEDIOL, 8-chloro-1,4-dimethyl-6-phenyl-4H-[1,2,4]triazolo[4,3-a][1,3,4]benzotriazepine, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Bracher, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-10-11 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Benzodiazepines and benzotriazepines as protein interaction inhibitors targeting bromodomains of the BET family.
Bioorg.Med.Chem., 20, 2012
|
|
4WA1
 
 | | The crystal structure of hemagglutinin from a H3N8 influenza virus isolated from New England harbor seals | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Yang, H, Villanueva, J.M, Gubareva, L.V, Stevens, J. | | Deposit date: | 2014-08-28 | | Release date: | 2015-01-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Structural and Functional Analysis of Surface Proteins from an A(H3N8) Influenza Virus Isolated from New England Harbor Seals.
J.Virol., 89, 2015
|
|
3GLB
 
 | | Crystal structure of the effector binding domain of a CATM variant (R156H) | | Descriptor: | (2Z,4Z)-HEXA-2,4-DIENEDIOIC ACID, GLYCEROL, HTH-type transcriptional regulator catM, ... | | Authors: | Ezezika, O.C, Craven, S.H, Neidle, E.L, Momany, C. | | Deposit date: | 2009-03-11 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Inducer responses of BenM, a LysR-type transcriptional regulator from Acinetobacter baylyi ADP1.
Mol.Microbiol., 72, 2009
|
|
3GLM
 
 | | Glutaconyl-coA decarboxylase A subunit from Clostridium symbiosum co-crystallized with crotonyl-coA | | Descriptor: | CHLORIDE ION, CROTONYL COENZYME A, Glutaconyl-CoA decarboxylase subunit A | | Authors: | Kress, D, Brugel, D, Buckel, W, Essen, L.-O. | | Deposit date: | 2009-03-12 | | Release date: | 2009-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An asymmetric model for Na+-translocating glutaconyl-CoA decarboxylases
J.Biol.Chem., 284, 2009
|
|
3U85
 
 | |
3U4X
 
 | | Crystal structure of a lectin from Camptosema pedicellatum seeds in complex with 5-bromo-4-chloro-3-indolyl-alpha-D-mannose | | Descriptor: | 5-bromo-4-chloro-1H-indol-3-yl alpha-D-mannopyranoside, CALCIUM ION, Camptosema pedicellatum lectin (CPL), ... | | Authors: | Rocha, B.A.M, Teixeira, C.S, Moura, T.R, Silva, H.C, Pereira-Junior, F.N, Nagano, C.S, Delatorre, P, Cavada, B.S. | | Deposit date: | 2011-10-10 | | Release date: | 2012-09-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of the lectin of Camptosema pedicellatum: implications of a conservative substitution at the hydrophobic subsite.
J.Biochem., 152, 2012
|
|
7LRN
 
 | |
3GMG
 
 | | Crystal structure of an uncharacterized conserved protein from Mycobacterium tuberculosis | | Descriptor: | Uncharacterized protein Rv1825/MT1873 | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Chang, S, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-13 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of an uncharacterized conserved protein from Mycobacterium tuberculosis
To be Published
|
|
4WEI
 
 | | Crystal structure of the F4 fimbrial adhesin FaeG in complex with lactose | | Descriptor: | K88 fimbrial protein AD, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Moonens, K, Van den Broeck, I, De Kerpel, M, Deboeck, F, Raymaekers, H, Remaut, H, De Greve, H. | | Deposit date: | 2014-09-10 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Functional Insight into the Carbohydrate Receptor Binding of F4 Fimbriae-producing Enterotoxigenic Escherichia coli.
J.Biol.Chem., 290, 2015
|
|
5WT9
 
 | | Complex structure of PD-1 and nivolumab-Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of Nivolumab, Light Chain of Nivolumab, ... | | Authors: | Tan, S, Zhang, H, Chai, Y, Song, H, Tong, Z, Wang, Q, Qi, J, Wong, G, Zhu, X, Liu, W.J, Gao, S, Wang, Z, Shi, Y, Yang, F, Gao, G.F, Yan, J. | | Deposit date: | 2016-12-10 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | An unexpected N-terminal loop in PD-1 dominates binding by nivolumab.
Nat Commun, 8, 2017
|
|
3GYD
 
 | |
4UTC
 
 | | Crystal structure of dengue 2 virus envelope glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ENVELOPE GLYCOPROTEIN E, ... | | Authors: | Kikuti, C, Rouvinski, A, Guardado-Calvo, P, Barba-Spaeth, G, Duquerroy, S, Vaney, M.C, Rey, F.A. | | Deposit date: | 2014-07-18 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Recognition Determinants of Broadly Neutralizing Human Antibodies Against Dengue Viruses.
Nature, 520, 2015
|
|
3U51
 
 | | Src in complex with DNA-templated macrocyclic inhibitor MC1 | | Descriptor: | Proto-oncogene tyrosine-protein kinase Src, macrocyclic inhibitor MC1 | | Authors: | Seeliger, M.A, Liu, D.R, Georghiou, G, Kleiner, R.E, Pulkoski-Gross, M. | | Deposit date: | 2011-10-10 | | Release date: | 2012-02-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.241 Å) | | Cite: | Highly specific, bisubstrate-competitive Src inhibitors from DNA-templated macrocycles.
Nat.Chem.Biol., 8, 2012
|
|
3GYT
 
 | | Nuclear receptor DAF-12 from parasitic nematode Strongyloides stercoralis in complex with its physiological ligand dafachronic acid delta 4 | | Descriptor: | (14beta,17alpha,25R)-3-oxocholest-4-en-26-oic acid, Nuclear hormone receptor of the steroid/thyroid hormone receptors superfamily, SRC1 | | Authors: | Zhou, X.E, Wang, Z, Suino-Powell, K, Motola, D.L, Conneely, A, Ogata, C, Sharma, K.K, Auchus, R.J, Kliewer, S.A, Xu, H.E, Mangelsdorf, D.J. | | Deposit date: | 2009-04-05 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of the nuclear receptor DAF-12 as a therapeutic target in parasitic nematodes.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4UV7
 
 | | The complex structure of extracellular domain of EGFR and GC1118A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, EPIDERMAL GROWTH FACTOR RECEPTOR, ... | | Authors: | Yoo, J.H, Cho, H.S. | | Deposit date: | 2014-08-05 | | Release date: | 2015-10-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Gc1118, an Anti-Egfr Antibody with a Distinct Binding Epitope and Superior Inhibitory Activity Against High-Affinity Egfr Ligands.
Mol.Cancer Ther., 15, 2016
|
|
3GMS
 
 | |
7LZH
 
 | | Structure of the glutamate receptor-like channel AtGLR3.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Gangwar, S.P, Green, M.N, Sobolevsky, A.I. | | Deposit date: | 2021-03-09 | | Release date: | 2021-07-28 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structure of the Arabidopsis thaliana glutamate receptor-like channel GLR3.4.
Mol.Cell, 81, 2021
|
|
5WVH
 
 | |
3U6H
 
 | | Crystal structure of c-Met in complex with pyrazolone inhibitor 26 | | Descriptor: | Hepatocyte growth factor receptor, N-{4-[(6,7-dimethoxyquinolin-4-yl)oxy]-3-fluorophenyl}-1,5-dimethyl-3-oxo-2-phenyl-2,3-dihydro-1H-pyrazole-4-carboxamide | | Authors: | Bellon, S.F, Whittington, D.A, Long, A.L. | | Deposit date: | 2011-10-12 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of novel class II c-Met inhibitors: 1. Identification of pyrazolone-based derivatives.
J.Med.Chem., 55, 2012
|
|
