6UYO
 
 | | Crystal structure of K37-acetylated SUMO1 in complex with PML-SIM | | Descriptor: | Protein PML, Small ubiquitin-related modifier 1 | | Authors: | Wahba, H.M, Gagnon, C, Mascle, X.H, Lussier-Price, M, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2019-11-14 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.639 Å) | | Cite: | Acetylation of SUMO1 Alters Interactions with the SIMs of PML and Daxx in a Protein-Specific Manner.
Structure, 28, 2020
|
|
7MH5
 
 | | Crystal structure of R. sphaeroides Photosynthetic Reaction Center variant; Y(M210)3-iodotyrosine | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Mathews, I, Weaver, J, Boxer, S.G. | | Deposit date: | 2021-04-14 | | Release date: | 2021-12-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Photosynthetic reaction center variants made via genetic code expansion show Tyr at M210 tunes the initial electron transfer mechanism.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6MBD
 
 | | Human Mcl-1 in complex with the designed peptide dM1 | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION, dM1 | | Authors: | Jenson, J.M, Keating, A.E. | | Deposit date: | 2018-08-29 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Tertiary Structural Motif Sequence Statistics Enable Facile Prediction and Design of Peptides that Bind Anti-apoptotic Bfl-1 and Mcl-1.
Structure, 27, 2019
|
|
8DRP
 
 | |
6P3Y
 
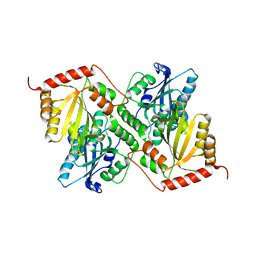 | | Crystal Structure of Full Length APOBEC3G E/Q (pH 7.4) | | Descriptor: | Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G, ZINC ION | | Authors: | Yang, H.J, Li, S.X, Chen, X.S. | | Deposit date: | 2019-05-25 | | Release date: | 2020-02-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Understanding the structural basis of HIV-1 restriction by the full length double-domain APOBEC3G.
Nat Commun, 11, 2020
|
|
6MBO
 
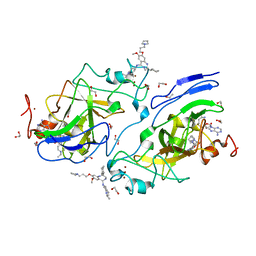 | | GLP Methyltransferase with Inhibitor EML741-P212121 Crystal Form | | Descriptor: | 1,2-ETHANEDIOL, 2-cyclohexyl-7-methoxy-N-[1-(propan-2-yl)piperidin-4-yl]-8-[3-(pyrrolidin-1-yl)propoxy]-3H-1,4-benzodiazepin-5-amine, Histone-lysine N-methyltransferase EHMT1, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2018-08-30 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Discovery of a Novel Chemotype of Histone Lysine Methyltransferase EHMT1/2 (GLP/G9a) Inhibitors: Rational Design, Synthesis, Biological Evaluation, and Co-crystal Structure.
J. Med. Chem., 62, 2019
|
|
6UYX
 
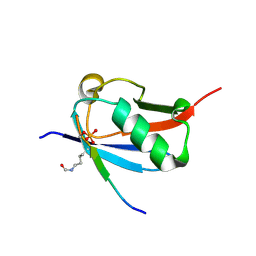 | | Crystal structure of K37-acetylated SUMO1 in complex with phosphorylated DAXX | | Descriptor: | Small ubiquitin-related modifier 1, phosphorylated DAXX | | Authors: | Wahba, H.M, Gagnon, C, Mascle, X.H, Lussier-Price, M, Cappadocia, L, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2019-11-14 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Acetylation of SUMO1 Alters Interactions with the SIMs of PML and Daxx in a Protein-Specific Manner.
Structure, 28, 2020
|
|
7MH4
 
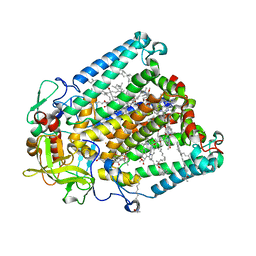 | | Crystal structure of R. sphaeroides Photosynthetic Reaction Center variant; Y(M210)3-bromotyrosine | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Mathews, I, Weaver, J, Boxer, S.G. | | Deposit date: | 2021-04-14 | | Release date: | 2021-12-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Photosynthetic reaction center variants made via genetic code expansion show Tyr at M210 tunes the initial electron transfer mechanism.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7A1D
 
 | | Cryo-EM map of the large glutamate dehydrogenase composed of 180 kDa subunits from Mycobacterium smegmatis (open conformation) | | Descriptor: | NAD-specific glutamate dehydrogenase | | Authors: | Lazaro, M, Melero, R, Huet, C, Lopez-Alonso, J.P, Delgado, S, Dodu, A, Bruch, E.M, Abriata, L.A, Alzari, P.M, Valle, M, Lisa, M.N. | | Deposit date: | 2020-08-12 | | Release date: | 2021-06-09 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.19 Å) | | Cite: | 3D architecture and structural flexibility revealed in the subfamily of large glutamate dehydrogenases by a mycobacterial enzyme.
Commun Biol, 4, 2021
|
|
8EA5
 
 | | NKG2D complexed with inhibitor 1a | | Descriptor: | (3S,5aS,8aR)-3-benzyl-6-[(3,5-dichlorophenyl)methyl]-1,4-dimethyloctahydropyrrolo[3,2-e][1,4]diazepine-2,5-dione, NKG2-D type II integral membrane protein, TRIETHYLENE GLYCOL | | Authors: | Thompson, A.A, Grant, J.C, Karpowich, N.K, Sharma, S. | | Deposit date: | 2022-08-28 | | Release date: | 2023-05-03 | | Last modified: | 2023-05-10 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Identification of small-molecule protein-protein interaction inhibitors for NKG2D.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6PGS
 
 | |
7MBN
 
 | | Cryo-EM structure of MLL1-NCP (H3K4M) complex, mode02 | | Descriptor: | DNA (146-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Park, S.H, Ayoub, A, Lee, Y.T, Dou, Y, Cho, U. | | Deposit date: | 2021-04-01 | | Release date: | 2021-12-29 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY | | Cite: | Regulation of MLL1 Methyltransferase Activity in Two Distinct Nucleosome Binding Modes.
Biochemistry, 61, 2022
|
|
8EA9
 
 | | NKG2D complexed with inhibitor 4d | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-[(1S)-1-[3,5-bis(trifluoromethyl)phenyl]-2-(dimethylamino)-2-oxoethyl]-4-[4-(trifluoromethyl)phenyl]pyridine-3-carboxamide, NKG2-D type II integral membrane protein, ... | | Authors: | Thompson, A.A, Grant, J.C, Karpowich, N.K, Sharma, S. | | Deposit date: | 2022-08-28 | | Release date: | 2023-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Identification of small-molecule protein-protein interaction inhibitors for NKG2D.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6ZW0
 
 | |
6UZ7
 
 | | K.lactis 80S ribosome with p/PE tRNA and eIF5B | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0, ... | | Authors: | Fernandez, I.S, Huang, B.Y. | | Deposit date: | 2019-11-14 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Long-range interdomain communications in eIF5B regulate GTP hydrolysis and translation initiation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6MC8
 
 | |
6UOL
 
 | |
7MT6
 
 | | Crystal structure of tryptophan synthase in complex with F9, Cs+, benzimidazole, pH7.8 - alpha aminoacrylate form - E(A-A)(BZI) | | Descriptor: | 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, BENZIMIDAZOLE, ... | | Authors: | Drago, V, Hilario, E, Dunn, M.F, Mueser, T.C, Mueller, L.J. | | Deposit date: | 2021-05-12 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Imaging active site chemistry and protonation states: NMR crystallography of the tryptophan synthase alpha-aminoacrylate intermediate.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6MC1
 
 | | Structure of MAP kinase phosphatase 5 in complex with 3,3-dimethyl-1-((9-(methylthio)-5,6-dihydrothieno[3,4-h]quinazolin-2-yl)thio)butan-2-one, an allosteric inhibitor | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 3,3-dimethyl-1-{[9-(methylsulfanyl)-5,6-dihydrothieno[3,4-h]quinazolin-2-yl]sulfanyl}butan-2-one, ACETATE ION, ... | | Authors: | Gannam, Z.T.K, Anderson, K.S, Bennett, A.M, Lolis, E. | | Deposit date: | 2018-08-30 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An allosteric site on MKP5 reveals a strategy for small-molecule inhibition.
Sci.Signal., 13, 2020
|
|
6ZVZ
 
 | |
6P46
 
 | | Cryo-EM structure of TMEM16F in digitonin with calcium bound | | Descriptor: | Anoctamin-6, CALCIUM ION | | Authors: | Feng, S, Dang, S, Han, T.W, Ye, W, Jin, P, Cheng, T, Li, J, Jan, Y.N, Jan, L.Y, Cheng, Y. | | Deposit date: | 2019-05-26 | | Release date: | 2019-07-24 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM Studies of TMEM16F Calcium-Activated Ion Channel Suggest Features Important for Lipid Scrambling.
Cell Rep, 28, 2019
|
|
8EAB
 
 | | NKG2D complexed with inhibitor 4f | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-[(1S)-2-oxo-1-[3-(trifluoromethyl)phenyl]-2-({4-[4-(trifluoromethyl)phenyl]pyridin-3-yl}amino)ethyl]-4-[4-(trifluoromethyl)phenyl]pyridine-3-carboxamide, NKG2-D type II integral membrane protein | | Authors: | Thompson, A.A, Grant, J.C, Karpowich, N.K, Sharma, S. | | Deposit date: | 2022-08-28 | | Release date: | 2023-05-03 | | Last modified: | 2023-05-10 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Identification of small-molecule protein-protein interaction inhibitors for NKG2D.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6P4N
 
 | | Salmonella typhi PltB Homopentamer with Neu5Ac-alpha-2-6-Gal-beta-1-4-GlcNAc Glycans | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Putative pertussis-like toxin subunit | | Authors: | Nguyen, T, Milano, S.K, Hillpot, E.C, Yang, Y.A, Song, J. | | Deposit date: | 2019-05-28 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Salmonella Typhoid Toxin PltB Subunit and Its Non-typhoidal Salmonella Ortholog Confer Differential Host Adaptation and Virulence.
Cell Host Microbe, 27, 2020
|
|
6UOX
 
 | | Structure of itraconazole-bound NPC1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(3-bromo-4-{4-[4-({(2R,4S)-2-(2,4-dichlorophenyl)-2-[(1H-1,2,4-triazol-1-yl)methyl]-1,3-dioxolan-4-yl}methoxy)phenyl]piperazin-1-yl}phenyl)-2-[(2S)-butan-2-yl]-2,4-dihydro-3H-1,2,4-triazol-3-one, ... | | Authors: | Long, T, Li, X. | | Deposit date: | 2019-10-15 | | Release date: | 2020-01-15 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.13 Å) | | Cite: | Structural basis for itraconazole-mediated NPC1 inhibition.
Nat Commun, 11, 2020
|
|
6ZOO
 
 | | Photosystem I reduced Plastocyanin Complex | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, ... | | Authors: | Nelson, N, Caspy, I, Shkolnisky, Y. | | Deposit date: | 2020-07-07 | | Release date: | 2021-06-16 | | Last modified: | 2021-07-07 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structure of plant photosystem I-plastocyanin complex reveals strong hydrophobic interactions.
Biochem.J., 478, 2021
|
|
