4GKI
 
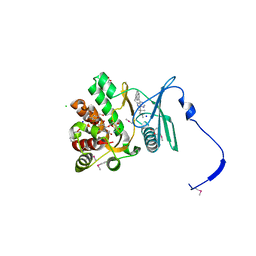 | | Crystal structure of the aminoglycoside phosphotransferase APH(3')-Ia, with substrate kanamycin and small molecule inhibitor 1-NM-PP1 | | Descriptor: | 1-tert-butyl-3-(naphthalen-1-ylmethyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, ACETATE ION, Aminoglycoside 3'-phosphotransferase AphA1-IAB, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Minasov, G, Egorova, O, Di Leo, R, Shakya, T, Spanogiannopoulos, P, Todorovic, N, Capretta, A, Wright, G.D, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-11 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure-guided optimization of protein kinase inhibitors reverses aminoglycoside antibiotic resistance.
Biochem.J., 454, 2013
|
|
4FMH
 
 | | Merkel Cell Polyomavirus VP1 in complex with Disialyllactose | | Descriptor: | CHLORIDE ION, GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose, ... | | Authors: | Neu, U, Hengel, H, Stehle, T. | | Deposit date: | 2012-06-17 | | Release date: | 2012-09-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of Merkel Cell Polyomavirus VP1 Complexes Define a Sialic Acid Binding Site Required for Infection.
Plos Pathog., 8, 2012
|
|
6FHW
 
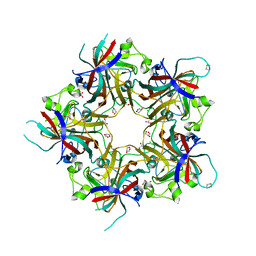
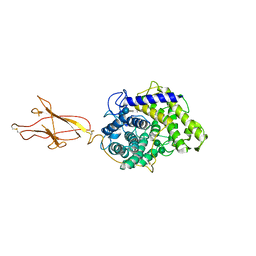 | | Structure of Hormoconis resinae Glucoamylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Roth, C, Moroz, O.V, Ariza, A, Friis, E.P, Davies, G.J, Wilson, K.S. | | Deposit date: | 2018-01-15 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural insight into industrially relevant glucoamylases: flexible positions of starch-binding domains.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
4FE1
 
 | | Improving the Accuracy of Macromolecular Structure Refinement at 7 A Resolution | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Fromme, R, Adams, P.D, Fromme, P, Levitt, M, Schroeder, G.F, Brunger, A.T. | | Deposit date: | 2012-05-29 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.9228 Å) | | Cite: | Improving the accuracy of macromolecular structure refinement at 7 A resolution.
Structure, 20, 2012
|
|
4GKH
 
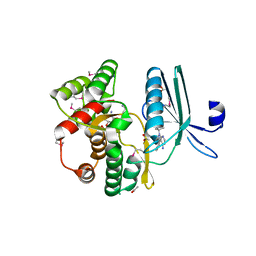 | | Crystal structure of the aminoglycoside phosphotransferase APH(3')-Ia, with substrate kanamycin and small molecule inhibitor 1-NA-PP1 | | Descriptor: | 1-tert-butyl-3-(naphthalen-1-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, ACETATE ION, Aminoglycoside 3'-phosphotransferase AphA1-IAB, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Minasov, G, Egorova, O, Di Leo, R, Shakya, T, Spanogiannopoulos, P, Todorovic, N, Capretta, A, Wright, G.D, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-11 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.863 Å) | | Cite: | Structure-guided optimization of protein kinase inhibitors reverses aminoglycoside antibiotic resistance.
Biochem.J., 454, 2013
|
|
4KML
 
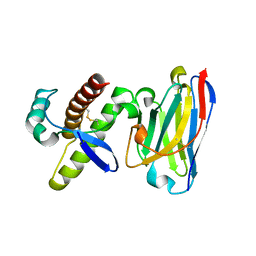 | | Probing the N-terminal beta-sheet conversion in the crystal structure of the full-length human prion protein bound to a Nanobody | | Descriptor: | Major prion protein, Nanobody | | Authors: | Abskharon, R.N.N, Giachin, G, Wohlkonig, A, Soror, S.H, Pardon, E, Legname, G, Steyaert, J. | | Deposit date: | 2013-05-08 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Probing the N-Terminal beta-Sheet Conversion in the Crystal Structure of the Human Prion Protein Bound to a Nanobody.
J.Am.Chem.Soc., 136, 2014
|
|
4TTG
 
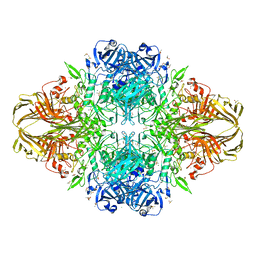 | | Beta-galactosidase (E. coli) in the presence of potassium chloride. | | Descriptor: | Beta-galactosidase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Juers, D.H. | | Deposit date: | 2014-06-20 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Elucidating factors important for monovalent cation selectivity in enzymes: E. coli beta-galactosidase as a model.
Phys Chem Chem Phys, 17, 2015
|
|
1ULV
 
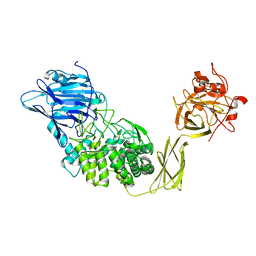 | | Crystal Structure of Glucodextranase Complexed with Acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, CALCIUM ION, glucodextranase | | Authors: | Mizuno, M, Tonozuka, T, Suzuki, S, Uotsu-Tomita, R, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2003-09-16 | | Release date: | 2003-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural insights into substrate specificity and function of glucodextranase
J.Biol.Chem., 279, 2004
|
|
3TOP
 
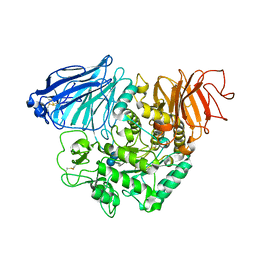 | | Crystral Structure of the C-terminal Subunit of Human Maltase-Glucoamylase in Complex with Acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Maltase-glucoamylase, intestinal | | Authors: | Shen, Y, Qin, X.H, Ren, L.M. | | Deposit date: | 2011-09-06 | | Release date: | 2011-11-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.881 Å) | | Cite: | Structural insight into substrate specificity of human intestinal maltase-glucoamylase
Protein Cell, 2, 2011
|
|
1X0R
 
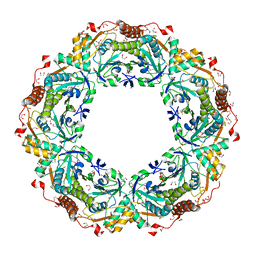 | | Thioredoxin Peroxidase from Aeropyrum pernix K1 | | Descriptor: | 1,2-ETHANEDIOL, Probable peroxiredoxin | | Authors: | Nakamura, T, Yamamoto, T, Inoue, T, Matsumura, H, Kobayashi, A, Hagihara, Y, Uegaki, K, Ataka, M, Kai, Y, Ishikawa, K. | | Deposit date: | 2005-03-28 | | Release date: | 2005-12-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of thioredoxin peroxidase from aerobic hyperthermophilic archaeon Aeropyrum pernix K1
Proteins, 62, 2006
|
|
1WPB
 
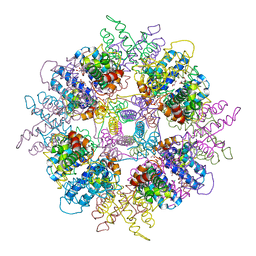 | | Structure of Escherichia coli yfbU gene product | | Descriptor: | CHLORIDE ION, GLYCEROL, hypothetical protein yfbU | | Authors: | Borek, D, Chen, Y, Zheng, M, Skarina, T, Savchenko, A, Edwards, A, Otwinowski, Z, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-09-01 | | Release date: | 2004-12-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Escherichia coli yfbU gene product
To be Published
|
|
5HCP
 
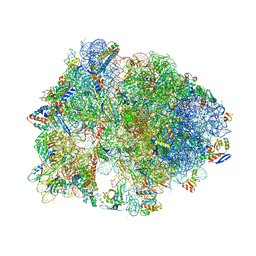 | | Crystal structure of antimicrobial peptide Metalnikowin bound to the Thermus thermophilus 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Gagnon, M.G, Roy, R.N, Lomakin, I.B, Florin, T, Mankin, A.S, Steitz, T.A. | | Deposit date: | 2016-01-04 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Structures of proline-rich peptides bound to the ribosome reveal a common mechanism of protein synthesis inhibition.
Nucleic Acids Res., 44, 2016
|
|
5HD1
 
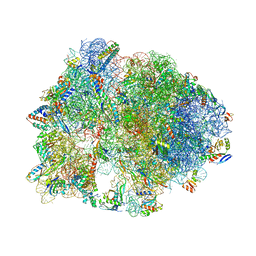 | | Crystal structure of antimicrobial peptide Pyrrhocoricin bound to the Thermus thermophilus 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Gagnon, M.G, Roy, R.N, Lomakin, I.B, Florin, T, Mankin, A.S, Steitz, T.A. | | Deposit date: | 2016-01-04 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of proline-rich peptides bound to the ribosome reveal a common mechanism of protein synthesis inhibition.
Nucleic Acids Res., 44, 2016
|
|
1RXS
 
 | | E. coli uridine phosphorylase: 2'-deoxyuridine phosphate complex | | Descriptor: | 2'-DEOXYURIDINE, META VANADATE, PHOSPHATE ION, ... | | Authors: | Caradoc-Davies, T.T, Cutfield, S.M, Lamont, I.L, Cutfield, J.F. | | Deposit date: | 2003-12-18 | | Release date: | 2004-04-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of escherichia coli uridine phosphorylase in two native and three complexed forms reveal basis of substrate specificity, induced conformational changes and influence of potassium
J.Mol.Biol., 337, 2004
|
|
1S72
 
 | | REFINED CRYSTAL STRUCTURE OF THE HALOARCULA MARISMORTUI LARGE RIBOSOMAL SUBUNIT AT 2.4 ANGSTROM RESOLUTION | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L10e, 50S ribosomal protein L11P, ... | | Authors: | Klein, D.J, Schmeing, T.M, Moore, P.B, Steitz, T.A. | | Deposit date: | 2004-01-28 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Roles of Ribosomal Proteins in the Structure, Assembly and Evolution of the Large Ribosomal Subunit
J.Mol.Biol., 340, 2004
|
|
1RWT
 
 | | Crystal Structure of Spinach Major Light-harvesting complex at 2.72 Angstrom Resolution | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Liu, Z, Yan, H, Wang, K, Kuang, T, Zhang, J, Gui, L, An, X, Chang, W. | | Deposit date: | 2003-12-17 | | Release date: | 2004-03-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Crystal structure of spinach major light-harvesting complex at 2.72 A resolution
Nature, 428, 2004
|
|
5HKV
 
 | | The crystal structure of the large ribosomal subunit of Staphylococcus aureus in complex with lincomycin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 23s ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Yonath, A, Matzov, D, Eyal, Z, Ben Hamou, R, Zimmerman, E, Rozenberg, H, Bashan, A, Fridman, M. | | Deposit date: | 2016-01-14 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.66 Å) | | Cite: | Structural insights of lincosamides targeting the ribosome of Staphylococcus aureus.
Nucleic Acids Res., 45, 2017
|
|
1S5L
 
 | | Architecture of the photosynthetic oxygen evolving center | | Descriptor: | 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, BETA-CAROTENE, BICARBONATE ION, ... | | Authors: | Ferreira, K.N, Iverson, T.M, Maghlaoui, K, Barber, J, Iwata, S. | | Deposit date: | 2004-01-21 | | Release date: | 2004-02-24 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Architecture of the Photosynthetic Oxygen-Evolving Center
Science, 303, 2004
|
|
5HUD
 
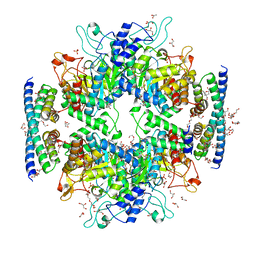 | | Non-covalent complex of and DAHP synthase and chorismate mutase from Corynebacterium glutamicum with bound transition state analog | | Descriptor: | 3-Deoxy-D-arabino-heptulosonate 7-phosphate (DAHP) synthase, 8-HYDROXY-2-OXA-BICYCLO[3.3.1]NON-6-ENE-3,5-DICARBOXYLIC ACID, Chorismate mutase, ... | | Authors: | Burschowsky, D, Heim, J.B, Thorbjoernsrud, H.V, Krengel, U. | | Deposit date: | 2016-01-27 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Inter-Enzyme Allosteric Regulation of Chorismate Mutase in Corynebacterium glutamicum: Structural Basis of Feedback Activation by Trp.
Biochemistry, 57, 2018
|
|
5IB8
 
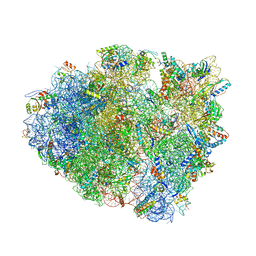 | | Structure of T. thermophilus 70S ribosome complex with mRNA, tRNAfMet and near-cognate tRNALys with U-G mismatch in the A-site | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2016-02-22 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | The ribosome prohibits the GU wobble geometry at the first position of the codon-anticodon helix.
Nucleic Acids Res., 44, 2016
|
|
1SMY
 
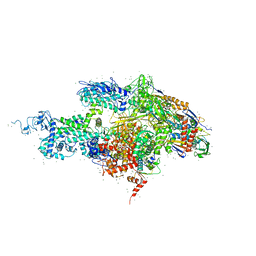 | | Structural basis for transcription regulation by alarmone ppGpp | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Artsimovitch, I, Patlan, V, Sekine, S, Vassylyeva, M.N, Hosaka, T, Ochi, K, Yokoyama, S, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-10 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for transcription regulation by alarmone ppGpp
Cell(Cambridge,Mass.), 117, 2004
|
|
5E24
 
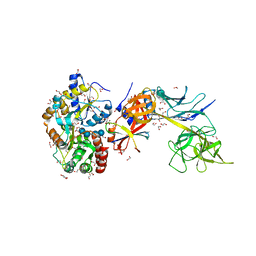 | | Structure of the Su(H)-Hairless-DNA Repressor Complex | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*AP*TP*CP*TP*TP*TP*CP*CP*CP*AP*CP*AP*GP*T)-3'), DNA (5'-D(*TP*TP*AP*CP*TP*GP*TP*GP*GP*GP*AP*AP*AP*GP*A)-3'), ... | | Authors: | Kovall, R.A, Yuan, Z. | | Deposit date: | 2015-09-30 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure and Function of the Su(H)-Hairless Repressor Complex, the Major Antagonist of Notch Signaling in Drosophila melanogaster.
Plos Biol., 14, 2016
|
|
6YQF
 
 | |
8QO1
 
 | | Asymmetric structure of the Borrelia bacteriophage BB1 procapsid, 3D class 3 | | Descriptor: | Cytosolic protein, DUF228 domain-containing protein, Decoration protein P03, ... | | Authors: | Rumnieks, J, Fuzik, T, Tars, K. | | Deposit date: | 2023-10-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (9.76 Å) | | Cite: | Structure of the Borrelia Bacteriophage phi BB1 Procapsid.
J.Mol.Biol., 435, 2023
|
|
7A4J
 
 | | Aquifex aeolicus lumazine synthase-derived nucleocapsid variant NC-4 | | Descriptor: | Antitermination protein N,6,7-dimethyl-8-ribityllumazine synthase,6,7-dimethyl-8-ribityllumazine synthase | | Authors: | Tetter, S, Hilvert, D. | | Deposit date: | 2020-08-19 | | Release date: | 2021-06-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Evolution of a virus-like architecture and packaging mechanism in a repurposed bacterial protein.
Science, 372, 2021
|
|
