8ZDX
 
 | | Crystal structure of MjHKU4r-CoV-1 RBD bound to hDPP4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Yang, M, Li, Z, Xu, Y, Zhang, S. | | Deposit date: | 2024-05-03 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of MjHKU4r-CoV-1 RBD bound to hDPP4
To Be Published
|
|
2DCM
 
 | | The Crystal Structure of S603A Mutated Prolyl Tripeptidyl Aminopeptidase Complexed with Substrate | | Descriptor: | GLYCYLALANYL-N-2-NAPHTHYL-L-PROLINEAMIDE, dipeptidyl aminopeptidase IV, putative | | Authors: | Nakajima, Y, Ito, K, Xu, Y, Yamada, N, Onohara, Y, Yoshimoto, T. | | Deposit date: | 2006-01-09 | | Release date: | 2006-09-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure and Mechanism of Tripeptidyl Activity of Prolyl Tripeptidyl Aminopeptidase from Porphyromonas gingivalis
J.Mol.Biol., 362, 2006
|
|
1U8E
 
 | | HUMAN DIPEPTIDYL PEPTIDASE IV/CD26 MUTANT Y547F | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bjelke, J.R, Christensen, J, Branner, S, Wagtmann, N, Olsen, C, Kanstrup, A.B, Rasmussen, H.B. | | Deposit date: | 2004-08-05 | | Release date: | 2004-08-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Tyrosine 547 constitutes an essential part of the catalytic mechanism of dipeptidyl peptidase IV
J.Biol.Chem., 279, 2004
|
|
4DTC
 
 | | Crystal Structure of DPP-IV with Compound C5 | | Descriptor: | Dipeptidyl peptidase 4, N-[(3R)-3-amino-4-(2,4,5-trifluorophenyl)butyl]-3'-(trifluoromethyl)biphenyl-4-carboxamide | | Authors: | Xiong, B, Zhu, L.R, Chen, D.Q, Zhao, Y.L, Jiang, F, Shen, J.K. | | Deposit date: | 2012-02-21 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of DPP-IV with Compound C5
To be Published
|
|
3NOX
 
 | | Crystal structure of human DPP-IV in complex with Sa-(+)-(6-(aminomethyl)-5-(2,4-dichlorophenyl)-7-methylimidazo[1,2-a]pyrimidin-2-yl)(morpholino)methanone | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl-peptidase 4 (CD26, ... | | Authors: | Klei, H.E. | | Deposit date: | 2010-06-25 | | Release date: | 2010-08-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.338 Å) | | Cite: | Discovery of 6-(Aminomethyl)-5-(2,4-dichlorophenyl)-7-methylimidazo[1,2-a]pyrimidine-2-carboxamides as Potent, Selective Dipeptidyl Peptidase-4 (DPP4) Inhibitors.
J.Med.Chem., 53, 2010
|
|
6L8Q
 
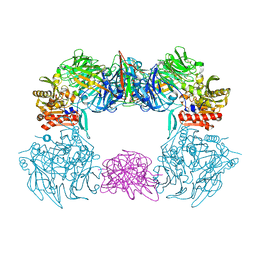 | | Complex structure of bat CD26 and MERS-RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Yuan, Y. | | Deposit date: | 2019-11-07 | | Release date: | 2019-12-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular Basis of Binding between Middle East Respiratory Syndrome Coronavirus and CD26 from Seven Bat Species.
J.Virol., 94, 2020
|
|
4QZV
 
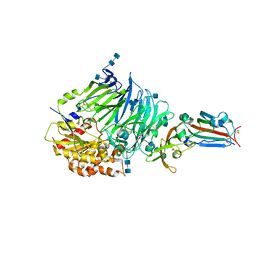 | | Bat-derived coronavirus HKU4 uses MERS-CoV receptor human CD26 for cell entry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Gao, F.G, Wang, Q.H, Qi, J.X, Lu, G.W. | | Deposit date: | 2014-07-29 | | Release date: | 2014-10-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.592 Å) | | Cite: | Bat Origins of MERS-CoV Supported by Bat Coronavirus HKU4 Usage of Human Receptor CD26.
Cell Host Microbe, 16, 2014
|
|
4WJL
 
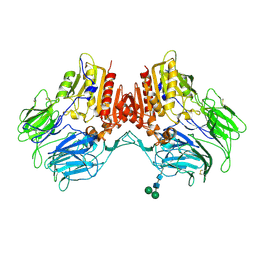 | | Structure of human dipeptidyl peptidase 10 (DPPY): a modulator of neuronal Kv4 channels | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Inactive dipeptidyl peptidase 10, ... | | Authors: | Bezerra, G.A, Dobrovetsky, E, Seitova, A, Fedosyuk, S, Dhe-Paganon, S, Gruber, K. | | Deposit date: | 2014-09-30 | | Release date: | 2015-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of human dipeptidyl peptidase 10 (DPPY): a modulator of neuronal Kv4 channels.
Sci Rep, 5, 2015
|
|
8WKU
 
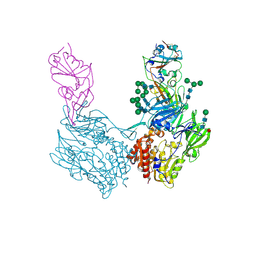 | | Complex structure of MjHKU4r-CoV-1 spike RBD bound to human CD26 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4 soluble form, MjHKU4r-CoV-1 spike RBD, ... | | Authors: | Zhao, Z.N, Chai, Y, Gao, F. | | Deposit date: | 2023-09-28 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular basis for receptor recognition and broad host tropism for merbecovirus MjHKU4r-CoV-1.
Embo Rep., 25, 2024
|
|
5OLJ
 
 | | Crystal structure of Porphyromonas gingivalis dipeptidyl peptidase 4 | | Descriptor: | Dipeptidyl peptidase IV, GLYCEROL | | Authors: | Fulop, V. | | Deposit date: | 2017-07-27 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Porphyromonas gingivalis dipeptidyl peptidase 4 and structure-activity relationships based on inhibitor profiling.
Eur J Med Chem, 139, 2017
|
|
7ZXS
 
 | | Crystal structure of DPP9 in complex with a 4-oxo-b-lactam based inhibitor, A295 | | Descriptor: | 1,2-ETHANEDIOL, 2-ethyl-2-methanoyl-~{N}-[3-[[4-(quinolin-8-ylmethyl)piperazin-1-yl]methyl]phenyl]butanamide, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ross, B, Huber, R. | | Deposit date: | 2022-05-22 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Chemoproteomics-Enabled Identification of 4-Oxo-beta-Lactams as Inhibitors of Dipeptidyl Peptidases 8 and 9.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
8PBA
 
 | |
1J2E
 
 | | Crystal structure of Human Dipeptidyl peptidase IV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase IV | | Authors: | Hiramatsu, H, Kyono, K, Higashiyama, Y, Fukushima, C, Shima, H, Sugiyama, S, Inaka, K, Yamamoto, A, Shimizu, R. | | Deposit date: | 2002-12-30 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure and function of human dipeptidyl peptidase IV, possessing a unique eight-bladed beta-propeller fold.
Biochem.Biophys.Res.Commun., 302, 2003
|
|
6HP8
 
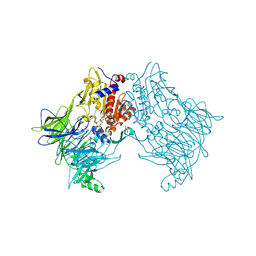 | | Crystal structure of DPP8 in complex with Val-BoroPro | | Descriptor: | Dipeptidyl peptidase 8, GLYCEROL, SODIUM ION, ... | | Authors: | Ross, B.H, Huber, R. | | Deposit date: | 2018-09-19 | | Release date: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Improvement of Protein Crystal Diffraction Using Post-Crystallization Methods: Infrared Laser Radiation Controls Crystal Order
Thesis, 2019
|
|
5VTA
 
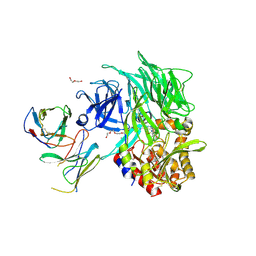 | | Co-Crystal Structure of DPPIV with a Chemibody Inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{4-[(3R)-3-amino-4-(2,4,5-trifluorophenyl)butanoyl]piperazin-1-yl}-N-(22-oxo-3,6,9,12,15,18-hexaoxa-21-azatricosan-1-yl)acetamide, ... | | Authors: | Wang, Z, Johnstone, S, Cheng, A. | | Deposit date: | 2017-05-16 | | Release date: | 2018-05-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-guided Discovery of Dual-recognition Chemibodies.
Sci Rep, 8, 2018
|
|
6QZV
 
 | |
3WQH
 
 | | Crystal Structure of human DPP-IV in complex with Anagliptin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, N-[2-({2-[(2S)-2-cyanopyrrolidin-1-yl]-2-oxoethyl}amino)-2-methylpropyl]-2-methylpyrazolo[1,5-a]pyrimidine-6-carboxamide | | Authors: | Watanabe, Y.S, Okada, S, Motoyama, T, Takahashi, R, Adachi, H, Oka, M. | | Deposit date: | 2014-01-27 | | Release date: | 2015-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Anagliptin, a potent dipeptidyl peptidase IV inhibitor: its single-crystal structure and enzyme interactions.
J Enzyme Inhib Med Chem, 30, 2015
|
|
7OR4
 
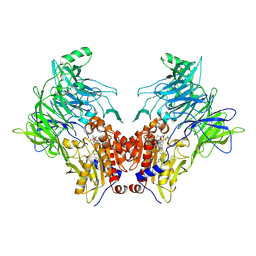 | | Crystal structure of DPP8 in complex with a 4-oxo-b-lactam based inhibitor, B142 | | Descriptor: | Dipeptidyl peptidase 8, methyl 3-[4-[(4-bromophenyl)methyl]piperazin-1-yl]carbonyl-5-[(2-ethyl-2-methanoyl-butanoyl)amino]benzoate, trimethylamine oxide | | Authors: | Ross, B, Huber, R. | | Deposit date: | 2021-06-04 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Chemoproteomics-Enabled Identification of 4-Oxo-beta-Lactams as Inhibitors of Dipeptidyl Peptidases 8 and 9.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7OZ7
 
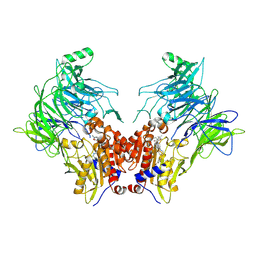 | | Crystal structure of DPP8 in complex with a 4-oxo-b-lactam based inhibitor, L84 | | Descriptor: | Dipeptidyl peptidase 8, trimethylamine oxide, ~{N}-[3-[[4-[(4-bromophenyl)methyl]piperazin-1-yl]methyl]phenyl]-2-ethyl-2-methanoyl-butanamide | | Authors: | Ross, B, Huber, R. | | Deposit date: | 2021-06-26 | | Release date: | 2021-07-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Chemoproteomics-Enabled Identification of 4-Oxo-beta-Lactams as Inhibitors of Dipeptidyl Peptidases 8 and 9.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
3KWF
 
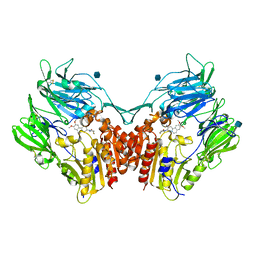 | | human DPP-IV with carmegliptin (S)-1-((2S,3S,11bS)-2-Amino-9,10-dimethoxy-1,3,4,6,7,11b-hexahydro-2H-pyrido[2,1-a]isoquinolin-3-yl)-4-fluoromethyl-pyrrolidin-2-one | | Descriptor: | (4S)-1-[(2S,3S,11bS)-2-amino-9,10-dimethoxy-1,3,4,6,7,11b-hexahydro-2H-pyrido[2,1-a]isoquinolin-3-yl]-4-(fluoromethyl)pyrrolidin-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4 | | Authors: | Hennig, M, Stihle, M, Thoma, R. | | Deposit date: | 2009-12-01 | | Release date: | 2010-01-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of carmegliptin: A potent and long-acting dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
6QZW
 
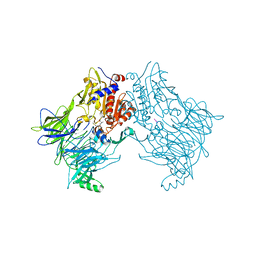 | |
4A5S
 
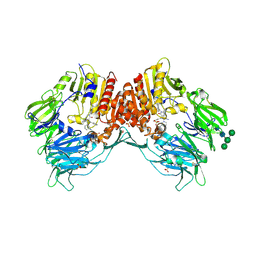 | | CRYSTAL STRUCTURE OF HUMAN DPP4 IN COMPLEX WITH A NOVAL HETEROCYCLIC DPP4 INHIBITOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-[(3S)-3-AMINOPIPERIDIN-1-YL]-5-BENZYL-4-OXO-3-(QUINOLIN-4-YLMETHYL)-4,5-DIHYDRO-3H-PYRROLO[3,2-D]PYRIMIDINE-7-CARBONITRILE, DIPEPTIDYL PEPTIDASE 4 SOLUBLE FORM, ... | | Authors: | Ostermann, N, Kroemer, M, Zink, F, Gerhartz, B, Sutton, J.M, Clark, D.E, Dunsdon, S.J, Fenton, G, Fillmore, A, Harris, N.V, Higgs, C, Hurley, C.A, Krintel, S.L, MacKenzie, R.E, Duttaroy, A, Gangl, E, Maniara, W, Sedrani, R, Namoto, K, Sirockin, F, Trappe, J, Hassiepen, U, Baeschlin, D.K. | | Deposit date: | 2011-10-28 | | Release date: | 2012-02-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Novel Heterocyclic Dpp-4 Inhibitors for the Treatment of Type 2 Diabetes.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
7JN7
 
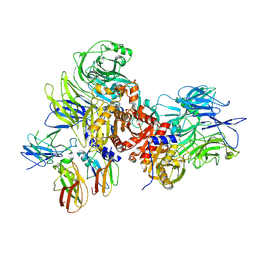 | | Human DPP9-CARD8 complex | | Descriptor: | Caspase recruitment domain-containing protein 8, Dipeptidyl peptidase 9, [(2~{R})-1-[(2~{R})-2-azanyl-3-methyl-butanoyl]pyrrolidin-2-yl]boronic acid | | Authors: | Sharif, H, Hollingsworth, L.R. | | Deposit date: | 2020-08-04 | | Release date: | 2021-05-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Dipeptidyl peptidase 9 sets a threshold for CARD8 inflammasome formation by sequestering its active C-terminal fragment.
Immunity, 54, 2021
|
|
7JKQ
 
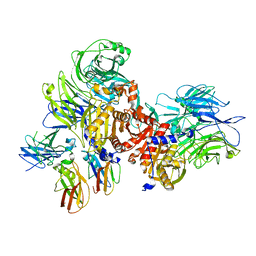 | | Human DPP9-CARD8 complex | | Descriptor: | Caspase recruitment domain-containing protein 8, Dipeptidyl peptidase 9 | | Authors: | Sharif, H, Hollingsworth, L.R. | | Deposit date: | 2020-07-28 | | Release date: | 2021-05-26 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Dipeptidyl peptidase 9 sets a threshold for CARD8 inflammasome formation by sequestering its active C-terminal fragment.
Immunity, 54, 2021
|
|
2Z3W
 
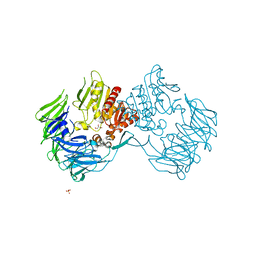 | | Prolyl tripeptidyl aminopeptidase mutant E636A | | Descriptor: | Dipeptidyl aminopeptidase IV, GLYCEROL, SULFATE ION | | Authors: | Xu, Y, Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2007-06-07 | | Release date: | 2008-02-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel inhibitor for prolyl tripeptidyl aminopeptidase from Porphyromonas gingivalis and details of substrate-recognition mechanism
J.Mol.Biol., 375, 2008
|
|
