1S0X
 
 | | Crystal structure of the human RORalpha ligand binding domain in complex with cholesterol sulfate at 2.2A | | Descriptor: | CHOLEST-5-EN-3-YL HYDROGEN SULFATE, Nuclear receptor ROR-alpha | | Authors: | Kallen, J, Schlaeppi, J.M, Bitsch, F, Delhon, I, Fournier, B. | | Deposit date: | 2004-01-05 | | Release date: | 2004-02-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the human RORalpha Ligand binding domain in complex with cholesterol sulfate at 2.2 A
J.Biol.Chem., 279, 2004
|
|
1S0Y
 
 | | The structure of trans-3-chloroacrylic acid dehalogenase, covalently inactivated by the mechanism-based inhibitor 3-bromopropiolate at 2.3 Angstrom resolution | | Descriptor: | MALONIC ACID, alpha-subunit of trans-3-chloroacrylic acid dehalogenase, beta-subunit of trans-3-chloroacrylic acid dehalogenase | | Authors: | de Jong, R.M, Brugman, W, Poelarends, G.J, Whitman, C.P, Dijkstra, B.W. | | Deposit date: | 2004-01-05 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The X-ray structure of trans-3-chloroacrylic acid dehalogenase reveals a novel hydration mechanism in the tautomerase superfamily
J.Biol.Chem., 279, 2004
|
|
1S0Z
 
 | | Crystal structure of the VDR LBD complexed to seocalcitol. | | Descriptor: | SEOCALCITOL, Vitamin D3 receptor | | Authors: | Tocchini-Valentini, G, Rochel, N, Wurtz, J.M, Moras, D. | | Deposit date: | 2004-01-05 | | Release date: | 2004-04-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the vitamin D nuclear receptor liganded with the vitamin D side chain analogues calcipotriol and seocalcitol, receptor agonists of clinical importance. Insights into a structural basis for the switching of calcipotriol to a receptor antagonist by further side chain modification.
J.Med.Chem., 47, 2004
|
|
1S10
 
 | | Snapshots of replication through an abasic lesion: structural basis for base substitution and frameshift | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*CP*TP*A)-3', 5'-D(*TP*CP*AP*GP*TP*AP*GP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Ling, H, Boudsocq, F, Woodgate, R, Yang, W. | | Deposit date: | 2004-01-05 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Snapshots of Replication through an Abasic Lesion; Structural Basis for Base Substitutions and Frameshifts.
Mol.Cell, 13, 2004
|
|
1S12
 
 | | Crystal structure of TM1457 | | Descriptor: | ACETATE ION, hypothetical protein TM1457 | | Authors: | Shin, D.H, Lou, Y, Jancarik, J, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-01-05 | | Release date: | 2004-12-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of TM1457 from Thermotoga maritima.
J.Struct.Biol., 152, 2005
|
|
1S13
 
 | | Human Heme Oxygenase Oxidatition of alpha- and gamma-meso-Phenylhemes | | Descriptor: | 2-PHENYLHEME, Heme oxygenase 1 | | Authors: | Wang, J, Niemevz, F, Lad, L, Buldain, G, Poulos, T.L, Ortiz de Montellano, P.R. | | Deposit date: | 2004-01-05 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Human heme oxygenase oxidation of 5- and 15-phenylhemes.
J.Biol.Chem., 279, 2004
|
|
1S14
 
 | | Crystal structure of Escherichia coli Topoisomerase IV ParE 24kDa subunit | | Descriptor: | NOVOBIOCIN, Topoisomerase IV subunit B | | Authors: | Wei, Y, Gross, C.H. | | Deposit date: | 2004-01-05 | | Release date: | 2004-05-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of Escherichia coli topoisomerase IV ParE subunit (24 and 43 kilodaltons): a single residue dictates differences in novobiocin potency against topoisomerase IV and DNA gyrase.
Antimicrob.Agents Chemother., 48, 2004
|
|
1S16
 
 | | Crystal Structure of E. coli Topoisomerase IV ParE 43kDa subunit complexed with ADPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SULFATE ION, ... | | Authors: | Wei, Y, Gross, C.H. | | Deposit date: | 2004-01-05 | | Release date: | 2004-05-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of Escherichia coli topoisomerase IV ParE subunit (24 and 43 kilodaltons): a single residue dictates differences in novobiocin potency against topoisomerase IV and DNA gyrase.
Antimicrob.Agents Chemother., 48, 2004
|
|
1S17
 
 | | Identification of Novel Potent Bicyclic Peptide Deformylase Inhibitors | | Descriptor: | 2-(3,4-DIHYDRO-3-OXO-2H-BENZO[B][1,4]THIAZIN-2-YL)-N-HYDROXYACETAMIDE, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Molteni, V, He, X, Nabakka, J, Yang, K, Kreusch, A, Gordon, P, Bursulaya, B, Ryder, N.S, Goldberg, R, He, Y. | | Deposit date: | 2004-01-05 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of novel potent bicyclic peptide deformylase inhibitors
Bioorg.Med.Chem.Lett., 14, 2004
|
|
1S18
 
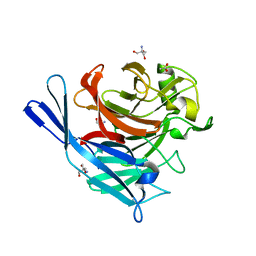 | | Structure and protein design of human apyrase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Dai, J, Liu, J, Deng, Y, Smith, T.M, Lu, M. | | Deposit date: | 2004-01-05 | | Release date: | 2004-03-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and protein design of a human platelet function inhibitor.
Cell(Cambridge,Mass.), 116, 2004
|
|
1S19
 
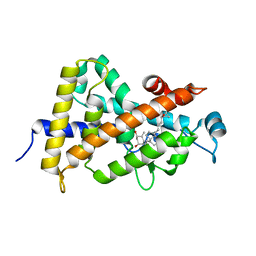 | | Crystal structure of VDR ligand binding domain complexed to calcipotriol. | | Descriptor: | CALCIPOTRIOL, Vitamin D3 receptor | | Authors: | Tocchini-Valentini, G, Rochel, N, Wurtz, J.M, Moras, D. | | Deposit date: | 2004-01-06 | | Release date: | 2004-04-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the vitamin D nuclear receptor liganded with the vitamin D side chain analogues calcipotriol and seocalcitol, receptor agonists of clinical importance. Insights into a structural basis for the switching of calcipotriol to a receptor antagonist by further side chain modification.
J.Med.Chem., 47, 2004
|
|
1S1A
 
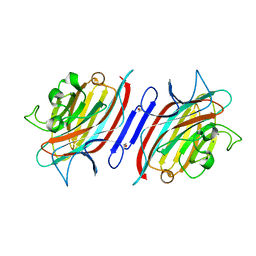 | | Pterocarpus angolensis seed lectin (PAL) with one binding site free and one binding site containing the disaccharide Man(a1-3)ManMe | | Descriptor: | CALCIUM ION, Lectin, MANGANESE (II) ION, ... | | Authors: | Buts, L, Imberty, A, Wyns, L, Beeckmans, S, Loris, R. | | Deposit date: | 2004-01-06 | | Release date: | 2005-01-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: |
|
|
1S1C
 
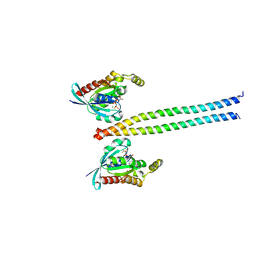 | | Crystal structure of the complex between the human RhoA and Rho-binding domain of human ROCKI | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Rho-associated, ... | | Authors: | Dvorsky, R, Blumenstein, L, Vetter, I.R, Ahmadian, M.R. | | Deposit date: | 2004-01-06 | | Release date: | 2004-02-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights into the Interaction of ROCKI with the Switch Regions of RhoA.
J.Biol.Chem., 279, 2004
|
|
1S1D
 
 | | Structure and protein design of human apyrase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Dai, J, Liu, J, Deng, Y, Smith, T.M, Lu, M. | | Deposit date: | 2004-01-06 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and protein design of a human platelet function inhibitor.
Cell(Cambridge,Mass.), 116, 2004
|
|
1S1E
 
 | | Crystal Structure of Kv Channel-interacting protein 1 (KChIP-1) | | Descriptor: | CALCIUM ION, Kv channel interacting protein 1 | | Authors: | Scannevin, R.H, Wang, K.-W, Jow, F, Megules, J, Kopsco, D.C, Edris, W, Carroll, K.C, Lu, Q, Xu, W.-X, Xu, Z.-B, Katz, A.H, Olland, S, Lin, L, Taylor, M, Stahl, M, Malakian, K, Somers, W, Mosyak, L, Bowlby, M.R, Chanda, P, Rhodes, K.J. | | Deposit date: | 2004-01-06 | | Release date: | 2005-01-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Two N-terminal domains of Kv4 K(+) channels regulate binding to and modulation by KChIP1.
Neuron, 41, 2004
|
|
1S1F
 
 | | Crystal Structure of Streptomyces Coelicolor A3(2) CYP158A2 from antibiotic biosynthetic pathways | | Descriptor: | 4-PHENYL-1H-IMIDAZOLE, GLYCEROL, MALONIC ACID, ... | | Authors: | Zhao, B, Lamb, D.C, Lei, L, Sundaramoorthy, M, Podust, L.M, Waterman, M.R. | | Deposit date: | 2004-01-06 | | Release date: | 2005-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding of Two Flaviolin Substrate Molecules, Oxidative Coupling, and Crystal Structure of Streptomyces coelicolor A3(2) Cytochrome P450 158A2
J.Biol.Chem., 280, 2005
|
|
1S1G
 
 | | Crystal Structure of Kv4.3 T1 Domain | | Descriptor: | Potassium voltage-gated channel subfamily D member 3, ZINC ION | | Authors: | Scannevin, R.H, Wang, K.W, Jow, F, Megules, J, Kopsco, D.C, Edris, W, Carroll, K.C, Lu, Q, Xu, W.X, Xu, Z.B, Katz, A.H, Olland, S, Lin, L, Taylor, M, Stahl, M, Malakian, K, Somers, W, Mosyak, L, Bowlby, M.R, Chanda, P, Rhodes, K.J. | | Deposit date: | 2004-01-06 | | Release date: | 2004-03-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Two N-terminal domains of Kv4 K(+) channels regulate binding to and modulation by KChIP1.
Neuron, 41, 2004
|
|
1S1J
 
 | | Crystal Structure of ZipA in complex with indoloquinolizin inhibitor 1 | | Descriptor: | (12bS)-1,2,3,4,12,12b-hexahydroindolo[2,3-a]quinolizin-7(6H)-one, Cell division protein zipA | | Authors: | Jenning, L.D, Foreman, K.W, Rush III, T.S, Tsao, D.H, Mosyak, L, Li, Y, Sukhdeo, M.N, Ding, W, Dushin, E.G, Kenney, C.H, Moghazeh, S.L, Peterson, P.J, Ruzin, A.V, Tuckman, M, Sutherland, A.G. | | Deposit date: | 2004-01-06 | | Release date: | 2004-05-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Design and synthesis of indolo[2,3-a]quinolizin-7-one inhibitors of the ZipA-FtsZ interaction
Bioorg.Med.Chem.Lett., 14, 2004
|
|
1S1K
 
 | |
1S1L
 
 | |
1S1M
 
 | | Crystal Structure of E. Coli CTP Synthetase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CTP synthase, IODIDE ION, ... | | Authors: | Endrizzi, J.A, Kim, H, Anderson, P.M, Baldwin, E.P. | | Deposit date: | 2004-01-06 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Escherichia coli Cytidine Triphosphate Synthetase, a Nucleotide-Regulated Glutamine Amidotransferase/ATP-Dependent Amidoligase Fusion Protein and Homologue of Anticancer and Antiparasitic Drug Targets
Biochemistry, 43, 2004
|
|
1S1N
 
 | | SH3 domain of human nephrocystin | | Descriptor: | Nephrocystin 1 | | Authors: | Le Maire, A, Weber, T, Saunier, S, Antignac, C, Ducruix, A, Dardel, F. | | Deposit date: | 2004-01-07 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the SH3 domain of human nephrocystin and analysis of a mutation-causing juvenile nephronophthisis.
Proteins, 59, 2005
|
|
1S1O
 
 | |
1S1P
 
 | | Crystal structures of prostaglandin D2 11-ketoreductase (AKR1C3) in complex with the non-steroidal anti-inflammatory drugs flufenamic acid and indomethacin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Aldo-keto reductase family 1 member C3, ... | | Authors: | Lovering, A.L, Ride, J.P, Bunce, C.M, Desmond, J.C, Cummings, S.M, White, S.A. | | Deposit date: | 2004-01-07 | | Release date: | 2004-03-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structures of prostaglandin D(2) 11-ketoreductase (AKR1C3) in complex with the nonsteroidal anti-inflammatory drugs flufenamic acid and indomethacin.
Cancer Res., 64, 2004
|
|
1S1Q
 
 | | TSG101(UEV) domain in complex with Ubiquitin | | Descriptor: | ACETIC ACID, COPPER (II) ION, SULFATE ION, ... | | Authors: | Sundquist, W.I, Schubert, H.L, Kelly, B.N, Hill, G.C, Holton, J.M, Hill, C.P. | | Deposit date: | 2004-01-07 | | Release date: | 2004-05-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ubiquitin recognition by the human TSG101 protein
Mol.Cell, 13, 2004
|
|
