1QAX
 
 | | TERNARY COMPLEX OF PSEUDOMONAS MEVALONII HMG-COA REDUCTASE WITH HMG-COA AND NAD+ | | Descriptor: | 3-HYDROXY-3-METHYLGLUTARYL-COENZYME A, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROTEIN (3-HYDROXY-3-METHYLGLUTARYL-COENZYME A REDUCTASE) | | Authors: | Tabernero, L, Bochar, D.A, Rodwell, V.W, Stauffacher, C.V. | | Deposit date: | 1999-04-06 | | Release date: | 1999-06-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Substrate-induced closure of the flap domain in the ternary complex structures provides insights into the mechanism of catalysis by 3-hydroxy-3-methylglutaryl-CoA reductase.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1QAY
 
 | | TERNARY COMPLEX OF PSEUDOMONAS MEVALONII HMG-COA REDUCTASE WITH MEVALONATE AND NAD+ | | Descriptor: | (R)-MEVALONATE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROTEIN (3-HYDROXY-3-METHYLGLUTARYL-COENZYME A REDUCTASE) | | Authors: | Tabernero, L, Bochar, D.A, Rodwell, V.W, Stauffacher, C.V. | | Deposit date: | 1999-04-07 | | Release date: | 1999-06-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Substrate-induced closure of the flap domain in the ternary complex structures provides insights into the mechanism of catalysis by 3-hydroxy-3-methylglutaryl-CoA reductase.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1QAZ
 
 | |
1QB0
 
 | | HUMAN CDC25B CATALYTIC DOMAIN | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, PROTEIN (M-PHASE INDUCER PHOSPHATASE 2 (CDC25B)), ... | | Authors: | Watenpaugh, K.D, Reynolds, R.A. | | Deposit date: | 1999-04-29 | | Release date: | 2000-04-29 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of the catalytic subunit of Cdc25B required for G2/M phase transition of the cell cycle.
J.Mol.Biol., 293, 1999
|
|
1QB1
 
 | |
1QB2
 
 | | CRYSTAL STRUCTURE OF THE CONSERVED SUBDOMAIN OF HUMAN PROTEIN SRP54M AT 2.1A RESOLUTION: EVIDENCE FOR THE MECHANISM OF SIGNAL PEPTIDE BINDING | | Descriptor: | HUMAN SIGNAL RECOGNITION PARTICLE 54 KD PROTEIN | | Authors: | Clemons Jr, W.M, Gowda, K, Black, S.D, Zwieb, C, Ramakrishnan, V. | | Deposit date: | 1999-04-29 | | Release date: | 1999-10-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the conserved subdomain of human protein SRP54M at 2.1 A resolution: evidence for the mechanism of signal peptide binding.
J.Mol.Biol., 292, 1999
|
|
1QB3
 
 | | CRYSTAL STRUCTURE OF THE CELL CYCLE REGULATORY PROTEIN CKS1 | | Descriptor: | CYCLIN-DEPENDENT KINASES REGULATORY SUBUNIT | | Authors: | Bourne, Y, Watson, M.H, Arvai, A.S, Bernstein, S.L, Reed, S.I, Tainer, J.A. | | Deposit date: | 1999-04-30 | | Release date: | 2000-08-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure and mutational analysis of the Saccharomyces cerevisiae cell cycle regulatory protein Cks1: implications for domain swapping, anion binding and protein interactions.
Structure Fold.Des., 8, 2000
|
|
1QB4
 
 | | CRYSTAL STRUCTURE OF MN(2+)-BOUND PHOSPHOENOLPYRUVATE CARBOXYLASE | | Descriptor: | ASPARTIC ACID, MANGANESE (II) ION, PHOSPHOENOLPYRUVATE CARBOXYLASE | | Authors: | Matsumura, H, Terada, M, Shirakata, S, Inoue, T, Yoshinaga, T, Izui, K, Kai, Y. | | Deposit date: | 1999-04-30 | | Release date: | 2002-05-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Plausible phosphoenolpyruvate binding site revealed by 2.6 A structure of Mn2+-bound phosphoenolpyruvate carboxylase from Escherichia coli
FEBS Lett., 458, 1999
|
|
1QB5
 
 | |
1QB6
 
 | | BOVINE TRYPSIN 3,3'-[3,5-DIFLUORO-4-METHYL-2, 6-PYRIDINEDIYLBIS(OXY)]BIS(BENZENECARBOXIMIDAMIDE) (ZK-805623) COMPLEX | | Descriptor: | 3,3'-[3,5-DIFLUORO-4-METHYL-2,6-PYRIDYLENEBIS(OXY)]-BIS(BENZENECARBOXIMIDAMIDE), CALCIUM ION, POTASSIUM ION, ... | | Authors: | Whitlow, M. | | Deposit date: | 1999-04-29 | | Release date: | 2000-04-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic analysis of potent and selective factor Xa inhibitors complexed to bovine trypsin.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1QB7
 
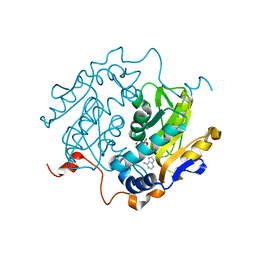 | | CRYSTAL STRUCTURES OF ADENINE PHOSPHORIBOSYLTRANSFERASE FROM LEISHMANIA DONOVANI. | | Descriptor: | ADENINE, ADENINE PHOSPHORIBOSYLTRANSFERASE, CITRIC ACID, ... | | Authors: | Phillips, C.L, Ullman, B, Brennan, R.G, Hill, C.P. | | Deposit date: | 1999-04-30 | | Release date: | 1999-07-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of adenine phosphoribosyltransferase from Leishmania donovani.
EMBO J., 18, 1999
|
|
1QB8
 
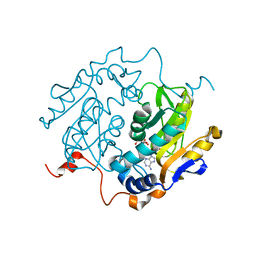 | |
1QB9
 
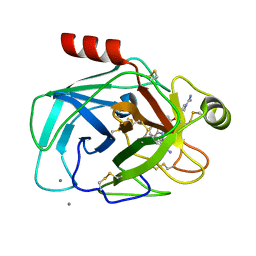 | |
1QBA
 
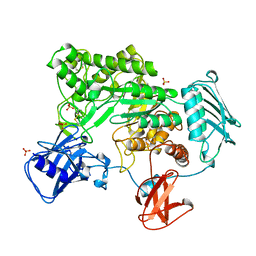 | | BACTERIAL CHITOBIASE, GLYCOSYL HYDROLASE FAMILY 20 | | Descriptor: | CHITOBIASE, SULFATE ION | | Authors: | Tews, I, Perrakis, A, Oppenheim, A, Dauter, Z, Wilson, K.S, Vorgias, C.E. | | Deposit date: | 1996-06-06 | | Release date: | 1997-01-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease.
Nat.Struct.Biol., 3, 1996
|
|
1QBB
 
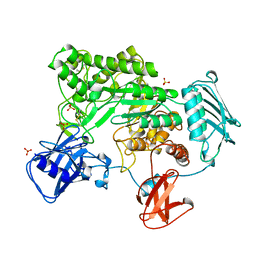 | | BACTERIAL CHITOBIASE COMPLEXED WITH CHITOBIOSE (DINAG) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITOBIASE, SULFATE ION | | Authors: | Tews, I, Perrakis, A, Oppenheim, A, Dauter, Z, Wilson, K.S, Vorgias, C.E. | | Deposit date: | 1996-06-07 | | Release date: | 1997-02-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease.
Nat.Struct.Biol., 3, 1996
|
|
1QBE
 
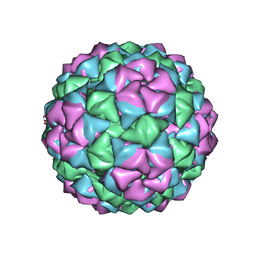 | | BACTERIOPHAGE Q BETA CAPSID | | Descriptor: | BACTERIOPHAGE Q BETA CAPSID | | Authors: | Liljas, L, Golmohammadi, R. | | Deposit date: | 1996-01-10 | | Release date: | 1996-07-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The crystal structure of bacteriophage Q beta at 3.5 A resolution.
Structure, 4, 1996
|
|
1QBF
 
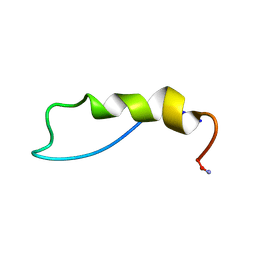 | |
1QBG
 
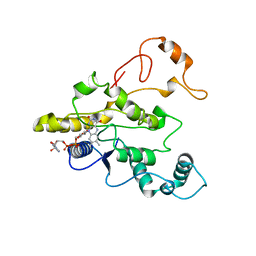 | | CRYSTAL STRUCTURE OF HUMAN DT-DIAPHORASE (NAD(P)H OXIDOREDUCTASE) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H DEHYDROGENASE [QUINONE] 1 | | Authors: | Skelly, J.V, Sanderson, M.R, Suter, D.A, Baumann, U, Gregory, D.S, Bennett, M, Hobbs, S.M, Neidle, S. | | Deposit date: | 1999-04-20 | | Release date: | 2000-04-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human DT-diaphorase: a model for interaction with the cytotoxic prodrug 5-(aziridin-1-yl)-2,4-dinitrobenzamide (CB1954).
J.Med.Chem., 42, 1999
|
|
1QBH
 
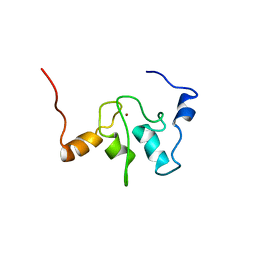 | | SOLUTION STRUCTURE OF A BACULOVIRAL INHIBITOR OF APOPTOSIS (IAP) REPEAT | | Descriptor: | INHIBITOR OF APOPTOSIS PROTEIN (2MIHB/C-IAP-1), ZINC ION | | Authors: | Hinds, M.G, Norton, R.S, Vaux, D.L, Day, C.L. | | Deposit date: | 1999-04-20 | | Release date: | 1999-10-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a baculoviral inhibitor of apoptosis (IAP) repeat.
Nat.Struct.Biol., 6, 1999
|
|
1QBI
 
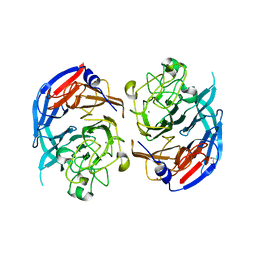 | | SOLUBLE QUINOPROTEIN GLUCOSE DEHYDROGENASE FROM ACINETOBACTER CALCOACETICUS | | Descriptor: | CALCIUM ION, GLYCEROL, PLATINUM (II) ION, ... | | Authors: | Oubrie, A, Rozeboom, H.J, Kalk, K.H, Duine, J.A, Dijkstra, B.W. | | Deposit date: | 1999-04-22 | | Release date: | 2000-02-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The 1.7 A crystal structure of the apo form of the soluble quinoprotein glucose dehydrogenase from Acinetobacter calcoaceticus reveals a novel internal conserved sequence repeat.
J.Mol.Biol., 289, 1999
|
|
1QBJ
 
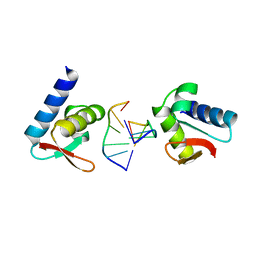 | | CRYSTAL STRUCTURE OF THE ZALPHA Z-DNA COMPLEX | | Descriptor: | DNA (5'-D(*TP*CP*GP*CP*GP*CP*G)-3'), PROTEIN (DOUBLE-STRANDED RNA SPECIFIC ADENOSINE DEAMINASE (ADAR1)) | | Authors: | Schwartz, T, Rould, M.A, Rich, A. | | Deposit date: | 1999-04-22 | | Release date: | 1999-07-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the Zalpha domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA.
Science, 284, 1999
|
|
1QBK
 
 | |
1QBL
 
 | |
1QBM
 
 | |
1QBN
 
 | |
