1Q34
 
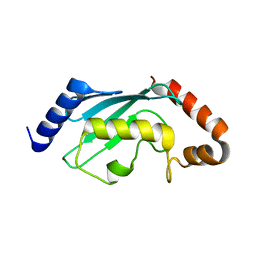 | | Crystal structures of two UBC (E2) enzymes of the ubiquitin-conjugating system in Caenorhabditis elegans | | Descriptor: | Ubiquitin-conjugating enzyme E2-21.5 kDa | | Authors: | Schormann, N, Lin, G, Li, S, Symersky, J, Qiu, S, Finley, J, Luo, D, Stanton, A, Carson, M, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-07-28 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of two UBC (E2) enzymes of the ubiquitin-conjugating system in Caenorhabditis elegans
To be Published
|
|
1Q35
 
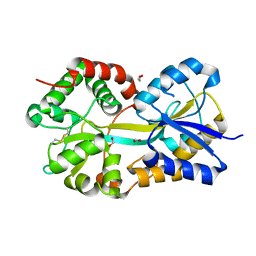 | | Crystal Structure of Pasteurella haemolytica Apo Ferric ion-Binding Protein A | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, iron binding protein FbpA | | Authors: | Shouldice, S.R, Dougan, D.R, Skene, R.J, Snell, G, Scheibe, D, Williams, P.A, Kirby, S, McRee, D.E, Schryvers, A.B, Tari, L.W. | | Deposit date: | 2003-07-28 | | Release date: | 2003-11-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of Pasteurella haemolytica ferric ion-binding protein A reveals a novel class of bacterial iron-binding proteins
J.Biol.Chem., 278, 2003
|
|
1Q36
 
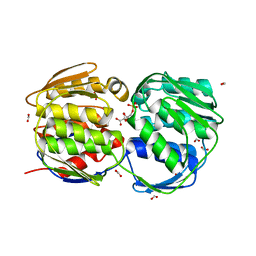 | | EPSP synthase (Asp313Ala) liganded with tetrahedral reaction intermediate | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, 5-(1-CARBOXY-1-PHOSPHONOOXY-ETHOXYL)-4-HYDROXY-3-PHOSPHONOOXY-CYCLOHEX-1-ENECARBOXYLIC ACID, FORMIC ACID | | Authors: | Eschenburg, S, Kabsch, W, Healy, M.L, Schonbrunn, E. | | Deposit date: | 2003-07-28 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A New View of the Mechanisms of UDP-N-Acetylglucosamine Enolpyruvyl Transferase (MurA) and 5-Enolpyruvylshikimate-3-phosphate Synthase (AroA) Derived from X-ray Structures of Their Tetrahedral Reaction Intermediate States.
J.Biol.Chem., 278, 2003
|
|
1Q38
 
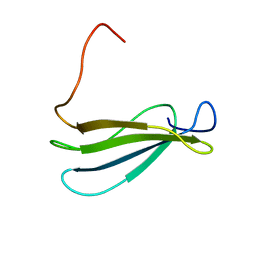 | | Anastellin | | Descriptor: | Fibronectin | | Authors: | Briknarova, K, Akerman, M.E, Hoyt, D.W, Ruoslahti, E, Ely, K.R. | | Deposit date: | 2003-07-28 | | Release date: | 2003-11-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Anastellin, an FN3 fragment with fibronectin polymerization activity, resembles amyloid fibril precursors
J.Mol.Biol., 332, 2003
|
|
1Q39
 
 | | Crystal structure of the DNA repair enzyme endonuclease-VIII (Nei) from E. coli: The WT enzyme at 2.8 resolution. | | Descriptor: | CALCIUM ION, Endonuclease VIII, ZINC ION | | Authors: | Golan, G, Zharkov, D.O, Feinberg, H, Fernandes, A.S, Zaika, E.I, Kycia, J.H, Grollman, A.P, Shoham, G. | | Deposit date: | 2003-07-29 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the uncomplexed DNA repair enzyme endonuclease VIII indicates significant interdomain flexibility.
Nucleic Acids Res., 33, 2005
|
|
1Q3A
 
 | | Crystal structure of the catalytic domain of human matrix metalloproteinase 10 | | Descriptor: | CALCIUM ION, N-ISOBUTYL-N-[4-METHOXYPHENYLSULFONYL]GLYCYL HYDROXAMIC ACID, Stromelysin-2, ... | | Authors: | Calderone, V, Bertini, I, Fragai, M, Luchinat, C, Mangani, S, Terni, B. | | Deposit date: | 2003-07-29 | | Release date: | 2004-04-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the catalytic domain of human matrix metalloproteinase 10.
J.Mol.Biol., 336, 2004
|
|
1Q3B
 
 | | Crystal structure of the DNA repair enzyme endonuclease-VIII (Nei) from E. coli: The R252A mutant at 2.05 resolution. | | Descriptor: | Endonuclease VIII, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Golan, G, Zharkov, D.O, Feinberg, H, Fernandes, A.S, Zaika, E.I, Kycia, J.H, Grollman, A.P, Shoham, G. | | Deposit date: | 2003-07-29 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the uncomplexed DNA repair enzyme endonuclease VIII indicates significant interdomain flexibility.
Nucleic Acids Res., 33, 2005
|
|
1Q3C
 
 | | Crystal structure of the DNA repair enzyme endonuclease-VIII (Nei) from E. coli: The E2A mutant at 2.3 resolution. | | Descriptor: | Endonuclease VIII, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Golan, G, Zharkov, D.O, Feinberg, H, Fernandes, A.S, Zaika, E.I, Kycia, J.H, Grollman, A.P, Shoham, G. | | Deposit date: | 2003-07-29 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the uncomplexed DNA repair enzyme endonuclease VIII indicates significant interdomain flexibility.
Nucleic Acids Res., 33, 2005
|
|
1Q3D
 
 | | GSK-3 Beta complexed with Staurosporine | | Descriptor: | GLYCOGEN SYNTHASE KINASE-3 BETA, STAUROSPORINE | | Authors: | Bertrand, J.A, Thieffine, S, Vulpetti, A, Cristiani, C, Valsasina, B, Knapp, S, Kalisz, H.M, Flocco, M. | | Deposit date: | 2003-07-29 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Characterization of the Gsk-3Beta Active Site Using Selective and Non-selective ATP-Mimetic Inhibitors
J.Mol.Biol., 333, 2003
|
|
1Q3E
 
 | | HCN2J 443-645 in the presence of cGMP | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 | | Authors: | Zagotta, W.N, Olivier, N.B, Black, K.D, Young, E.C, Olson, R, Gouaux, J.E. | | Deposit date: | 2003-07-29 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for modulation and agonist specificity of HCN pacemaker channels
Nature, 425, 2003
|
|
1Q3F
 
 | | Uracil DNA glycosylase bound to a cationic 1-aza-2'-deoxyribose-containing DNA | | Descriptor: | 5'-D(*AP*AP*AP*GP*AP*TP*AP*AP*CP*A)-3', 5'-D(*TP*GP*TP*(NRI)P*AP*TP*CP*TP*T)-3', PHOSPHATE ION, ... | | Authors: | Bianchet, M.A, Seiple, L.A, Jiang, Y.L, Ichikawa, Y, Amzel, L.M, Stivers, J.T. | | Deposit date: | 2003-07-29 | | Release date: | 2004-03-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Electrostatic guidance of glycosyl cation migration along the reaction coordinate of uracil DNA glycosylase.
Biochemistry, 42, 2003
|
|
1Q3G
 
 | | MurA (Asp305Ala) liganded with tetrahedral reaction intermediate | | Descriptor: | 1,2-ETHANEDIOL, 3'-1-CARBOXY-1-PHOSPHONOOXY-ETHOXY-URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Eschenburg, S, Kabsch, W, Healy, M.L, Schonbrunn, E. | | Deposit date: | 2003-07-29 | | Release date: | 2003-12-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A New View of the Mechanisms of UDP-N-Acetylglucosamine Enolpyruvyl Transferase (MurA) and 5-Enolpyruvylshikimate-3-phosphate Synthase (AroA) Derived from X-ray Structures of Their Tetrahedral Reaction Intermediate States.
J.Biol.Chem., 278, 2003
|
|
1Q3H
 
 | | mouse CFTR NBD1 with AMP.PNP | | Descriptor: | ACETIC ACID, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION, ... | | Authors: | Lewis, H.A, Buchanan, S.G, Burley, S.K, Conners, K, Dickey, M, Dorwart, M, Fowler, R, Gao, X, Guggino, W.B, Hendrickson, W.A. | | Deposit date: | 2003-07-29 | | Release date: | 2003-12-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of nucleotide-binding domain 1 of the cystic fibrosis transmembrane conductance regulator.
Embo J., 23, 2004
|
|
1Q3I
 
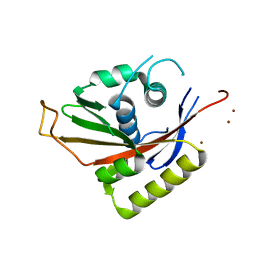 | | Crystal Structure of Na,K-ATPase N-domain | | Descriptor: | NICKEL (II) ION, Na,K-ATPase | | Authors: | Hakansson, K.O. | | Deposit date: | 2003-07-30 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Crystallographic Structure of Na,K-ATPase N-domain at 2.6 A Resolution
J.Mol.Biol., 332, 2003
|
|
1Q3J
 
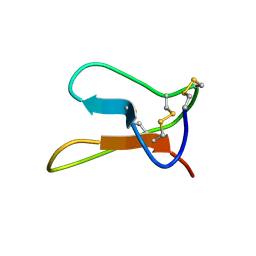 | | Solution structure of ALO3: a new knottin-type antifungal peptide from the insect Acrocinus longimanus | | Descriptor: | ALO3 | | Authors: | Barbault, F, Landon, C, Guenneugues, M, Meyer, J.P, Schott, V, Dimarrcq, J.L, Vovelle, F. | | Deposit date: | 2003-07-30 | | Release date: | 2003-12-23 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Alo-3: a new knottin-type antifungal peptide from the insect Acrocinus longimanus.
Biochemistry, 42, 2003
|
|
1Q3K
 
 | |
1Q3L
 
 | |
1Q3M
 
 | |
1Q3N
 
 | | Crystal structure of KDO8P synthase in its binary complex with substrate PEP | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, PHOSPHOENOLPYRUVATE | | Authors: | Vainer, R, Belakhov, V, Rabkin, E, Baasov, T, Adir, N. | | Deposit date: | 2003-07-31 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of Escherichia coli KDO8P synthase complexes reveal the source of catalytic irreversibility.
J.Mol.Biol., 351, 2005
|
|
1Q3O
 
 | | Crystal structure of the Shank PDZ-ligand complex reveals a class I PDZ interaction and a novel PDZ-PDZ dimerization | | Descriptor: | BROMIDE ION, Shank1 | | Authors: | Im, Y.J, Lee, J.H, Park, S.H, Park, S.J, Rho, S.-H, Kang, G.B, Kim, E, Eom, S.H. | | Deposit date: | 2003-07-31 | | Release date: | 2004-01-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Shank PDZ-ligand complex reveals a class I PDZ interaction and a novel PDZ-PDZ dimerization
J.Biol.Chem., 278, 2003
|
|
1Q3P
 
 | | Crystal structure of the Shank PDZ-ligand complex reveals a class I PDZ interaction and a novel PDZ-PDZ dimerization | | Descriptor: | C-terminal hexapeptide from Guanylate kinase-associated protein, Shank1 | | Authors: | Im, Y.J, Lee, J.H, Park, S.H, Park, S.J, Rho, S.-H, Kang, G.B, Kim, E, Eom, S.H. | | Deposit date: | 2003-07-31 | | Release date: | 2004-01-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the Shank PDZ-ligand complex reveals a class I PDZ interaction and a novel PDZ-PDZ dimerization
J.Biol.Chem., 278, 2003
|
|
1Q3Q
 
 | | Crystal structure of the chaperonin from Thermococcus strain KS-1 (two-point mutant complexed with AMP-PNP) | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Thermosome alpha subunit | | Authors: | Shomura, Y, Yoshida, T, Iizuka, R, Maruyama, T, Yohda, M, Miki, K. | | Deposit date: | 2003-07-31 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the Group II Chaperonin from Thermococcus strain KS-1: Steric Hindrance by the Substituted Amino Acid, and Inter-subunit Rearrangement between Two Crystal Forms.
J.Mol.Biol., 335, 2004
|
|
1Q3R
 
 | | Crystal structure of the chaperonin from Thermococcus strain KS-1 (nucleotide-free form of single mutant) | | Descriptor: | SULFATE ION, Thermosome alpha subunit | | Authors: | Shomura, Y, Yoshida, T, Iizuka, R, Maruyama, T, Yohda, M, Miki, K. | | Deposit date: | 2003-07-31 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structures of the Group II Chaperonin from Thermococcus strain KS-1: Steric Hindrance by the Substituted Amino Acid, and Inter-subunit Rearrangement between Two Crystal Forms.
J.Mol.Biol., 335, 2004
|
|
1Q3S
 
 | | Crystal structure of the chaperonin from Thermococcus strain KS-1 (FormIII crystal complexed with ADP) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Thermosome alpha subunit | | Authors: | Shomura, Y, Yoshida, T, Iizuka, R, Maruyama, T, Yohda, M, Miki, K. | | Deposit date: | 2003-07-31 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structures of the Group II Chaperonin from Thermococcus strain KS-1: Steric Hindrance by the Substituted Amino Acid, and Inter-subunit Rearrangement between Two Crystal Forms.
J.Mol.Biol., 335, 2004
|
|
1Q3T
 
 | | Solution structure and function of an essential CMP kinase of Streptococcus pneumoniae | | Descriptor: | Cytidylate kinase | | Authors: | Yu, L, Mack, J, Hajduk, P.J, Kakavas, S.J, Saiki, A.Y, Lerner, C.G, Olejniczak, E.T. | | Deposit date: | 2003-07-31 | | Release date: | 2004-08-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and function of an essential CMP kinase of Streptococcus pneumoniae
Protein Sci., 12, 2003
|
|
