1IE0
 
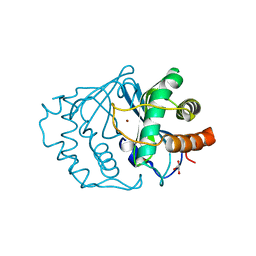 | | CRYSTAL STRUCTURE OF LUXS | | Descriptor: | AUTOINDUCER-2 PRODUCTION PROTEIN LUXS, GLYCEROL, ZINC ION | | Authors: | Hilgers, M.T, Ludwig, M.L. | | Deposit date: | 2001-04-05 | | Release date: | 2001-10-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the quorum-sensing protein LuxS reveals a catalytic metal site.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1IE1
 
 | | NMR Solution Structure of an In Vitro Selected RNA which is Sequence Specifically Recognized by Hamster Nucleolin RBD12. | | Descriptor: | 5'-R(*GP*GP*CP*CP*GP*AP*AP*AP*UP*CP*CP*CP*GP*AP*AP*GP*UP*AP*GP*GP*CP*C)-3' | | Authors: | Bouvet, P, Allain, F.H.-T, Finger, L.D, Dieckmann, T, Feigon, J. | | Deposit date: | 2001-04-05 | | Release date: | 2001-06-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recognition of pre-formed and flexible elements of an RNA stem-loop by nucleolin.
J.Mol.Biol., 309, 2001
|
|
1IE2
 
 | | Solution Structure of an In Vitro Selected RNA which is Sequence Specifically Recognized by RBD12 of Hamster Nucleolin.sNRE (anti) | | Descriptor: | 5'-R(*GP*GP*CP*CP*GP*AP*AP*AP*UP*CP*CP*CP*GP*AP*AP*GP*UP*AP*GP*GP*CP*C)-3' | | Authors: | Bouvet, P, Allain, F.H.-T, Finger, L.D, Dieckmann, T, Feigon, J. | | Deposit date: | 2001-04-05 | | Release date: | 2001-06-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Recognition of pre-formed and flexible elements of an RNA stem-loop by nucleolin.
J.Mol.Biol., 309, 2001
|
|
1IE3
 
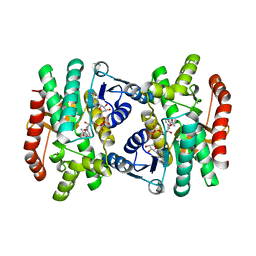 | | CRYSTAL STRUCTURE OF R153C E. COLI MALATE DEHYDROGENASE | | Descriptor: | MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PYRUVIC ACID | | Authors: | Bell, J.K, Yennawar, H.P, Wright, S.K, Thompson, J.R, Viola, R.E, Banaszak, L.J. | | Deposit date: | 2001-04-05 | | Release date: | 2001-09-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Analyses of a Malate Dehydrogenase with a Variable Active Site
J.Biol.Chem., 276, 2001
|
|
1IE4
 
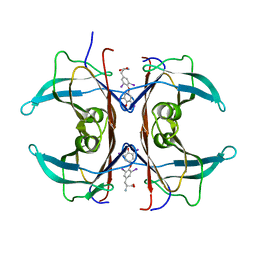 | | RAT TRANSTHYRETIN COMPLEX WITH THYROXINE (T4) | | Descriptor: | 3,5,3',5'-TETRAIODO-L-THYRONINE, TRANSTHYRETIN | | Authors: | Wojtczak, A. | | Deposit date: | 2001-04-06 | | Release date: | 2002-04-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of rat transthyretin (rTTR) complex with thyroxine at 2.5 A resolution: first non-biased insight into thyroxine binding reveals different hormone orientation in two binding sites.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1IE5
 
 | | NMR STRUCTURE OF THE THIRD IMMUNOGLOBULIN DOMAIN FROM THE NEURAL CELL ADHESION MOLECULE. | | Descriptor: | NEURAL CELL ADHESION MOLECULE | | Authors: | Atkins, A.R, Chung, J, Deechongkit, S, Little, E.B, Edelman, G.M, Wright, P.E, Cunningham, B.A, Dyson, H.J. | | Deposit date: | 2001-04-06 | | Release date: | 2001-08-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the third immunoglobulin domain of the neural cell adhesion molecule N-CAM: can solution studies define the mechanism of homophilic binding?
J.Mol.Biol., 311, 2001
|
|
1IE6
 
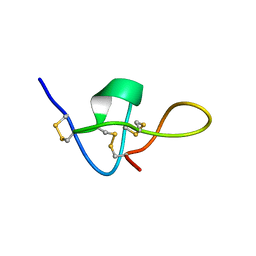 | | SOLUTION STRUCTURE OF IMPERATOXIN A | | Descriptor: | IMPERATOXIN A | | Authors: | Lee, C.W, Takeuchi, K, Takahashi, H, Sato, K, Shimada, I, Kim, D.H, Kim, J.I. | | Deposit date: | 2001-04-07 | | Release date: | 2003-06-10 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of the high-affinity activation of type 1 ryanodine receptors by imperatoxin A.
Biochem.J., 377, 2004
|
|
1IE7
 
 | | PHOSPHATE INHIBITED BACILLUS PASTEURII UREASE CRYSTAL STRUCTURE | | Descriptor: | NICKEL (II) ION, PHOSPHATE ION, UREASE ALPHA SUBUNIT, ... | | Authors: | Benini, S, Rypniewski, W.R, Wilson, K.S, Ciurli, S, Mangani, S. | | Deposit date: | 2001-04-09 | | Release date: | 2001-04-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-based rationalization of urease inhibition by phosphate: novel insights into the enzyme mechanism.
J.Biol.Inorg.Chem., 6, 2001
|
|
1IE8
 
 | | Crystal Structure Of The Nuclear Receptor For Vitamin D Ligand Binding Domain Bound to KH1060 | | Descriptor: | 5-(2-{1-[1-(4-ETHYL-4-HYDROXY-HEXYLOXY)-ETHYL]-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE}-ETHYLIDENE)-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, VITAMIN D3 RECEPTOR | | Authors: | Tocchini-Valentini, G, Rochel, N, Wurtz, J.M, Mitschler, A, Moras, D. | | Deposit date: | 2001-04-09 | | Release date: | 2001-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal structures of the vitamin D receptor complexed to superagonist 20-epi ligands.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1IE9
 
 | | Crystal Structure Of The Nuclear Receptor For Vitamin D Ligand Binding Domain Bound to MC1288 | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, VITAMIN D3 RECEPTOR | | Authors: | Tocchini-Valentini, G, Rochel, N, Wurtz, J.M, Mitschler, A, Moras, D. | | Deposit date: | 2001-04-09 | | Release date: | 2001-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structures of the vitamin D receptor complexed to superagonist 20-epi ligands.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1IEA
 
 | | HISTOCOMPATIBILITY ANTIGEN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MHC CLASS II I-EK | | Authors: | Fremont, D.H, Hendrickson, W.A, Marrack, P, Kappler, J. | | Deposit date: | 1996-04-05 | | Release date: | 1997-06-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of an MHC class II molecule with covalently bound single peptides.
Science, 272, 1996
|
|
1IEB
 
 | | HISTOCOMPATIBILITY ANTIGEN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MHC CLASS II I-EK, SULFATE ION | | Authors: | Fremont, D.H, Hendrickson, W.A, Marrack, P, Kappler, J. | | Deposit date: | 1996-04-05 | | Release date: | 1997-06-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of an MHC class II molecule with covalently bound single peptides.
Science, 272, 1996
|
|
1IEC
 
 | | CRYSTAL STRUCTURE OF THE CATALYTIC SITE MUTANT (H157A) OF THE HUMAN CYTOMEGALOVIRUS PROTEASE | | Descriptor: | CAPSID PROTEIN P40: ASSEMBLIN PROTEASE | | Authors: | Khayat, R, Batra, R, Massariol, M.J, Lagace, L, Tong, L. | | Deposit date: | 2001-04-09 | | Release date: | 2001-06-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Investigating the role of histidine 157 in the catalytic activity of human cytomegalovirus protease.
Biochemistry, 40, 2001
|
|
1IED
 
 | | CRYSTAL STRUCTURE OF THE CATALYTIC SITE MUTANT (H157E) OF THE HUMAN CYTOMEGALOVIRUS PROTEASE | | Descriptor: | CAPSID PROTEIN P40: ASSEMBLIN PROTEASE | | Authors: | Khayat, R, Batra, R, Massariol, M.J, Lagace, L, Tong, L. | | Deposit date: | 2001-04-09 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Investigating the role of histidine 157 in the catalytic activity of human cytomegalovirus protease.
Biochemistry, 40, 2001
|
|
1IEE
 
 | | STRUCTURE OF TETRAGONAL HEN EGG WHITE LYSOZYME AT 0.94 A FROM CRYSTALS GROWN BY THE COUNTER-DIFFUSION METHOD | | Descriptor: | CHLORIDE ION, LYSOZYME C, SODIUM ION | | Authors: | Sauter, C, Otalora, F, Gavira, J.-A, Vidal, O, Giege, R, Garcia-Ruiz, J.-M. | | Deposit date: | 2001-04-09 | | Release date: | 2001-08-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Structure of tetragonal hen egg-white lysozyme at 0.94 A from crystals grown by the counter-diffusion method.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1IEF
 
 | | CRYSTAL STRUCTURE OF THE CATALYTIC SITE MUTANT S134A OF THE HUMAN CYTOMEGALOVIRUS PROTEASE | | Descriptor: | CAPSID PROTEIN P40: ASSEMBLIN PROTEASE | | Authors: | Khayat, R, Batra, R, Massariol, M.J, Lagace, L, Tong, L. | | Deposit date: | 2001-04-09 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Investigating the role of histidine 157 in the catalytic activity of human cytomegalovirus protease.
Biochemistry, 40, 2001
|
|
1IEG
 
 | | CRYSTAL STRUCTURE OF THE CATALYTIC SITE MUTANT S134A/H157A OF THE HUMAN CYTOMEGALOVIRUS PROTEASE | | Descriptor: | CAPSID PROTEIN P40: ASSEMBLIN PROTEASE | | Authors: | Khayat, R, Batra, R, Massariol, M.J, Lagace, L, Tong, L. | | Deposit date: | 2001-04-09 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Investigating the role of histidine 157 in the catalytic activity of human cytomegalovirus protease.
Biochemistry, 40, 2001
|
|
1IEH
 
 | | SOLUTION STRUCTURE OF A SOLUBLE SINGLE-DOMAIN ANTIBODY WITH HYDROPHOBIC RESIDUES TYPICAL OF A VL/VH INTERFACE | | Descriptor: | BRUC.D4.4 | | Authors: | Vranken, W, Tolkatchev, D, Xu, P, Tanha, J, Chen, Z, Narang, S, Ni, F. | | Deposit date: | 2001-04-09 | | Release date: | 2002-08-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a llama single-domain antibody with hydrophobic residues typical of the VH/VL interface.
Biochemistry, 41, 2002
|
|
1IEI
 
 | | CRYSTAL STRUCTURE OF HUMAN ALDOSE REDUCTASE COMPLEXED WITH THE INHIBITOR ZENARESTAT. | | Descriptor: | ALDOSE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [3-(4-BROMO-2-FLUORO-BENZYL)-7-CHLORO-2,4-DIOXO-3,4-DIHYDRO-2H-QUINAZOLIN-1-YL]-ACETIC ACID | | Authors: | Kinoshita, T, Miyake, H, Fujii, T, Takakura, S, Goto, T. | | Deposit date: | 2001-04-09 | | Release date: | 2002-04-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of human recombinant aldose reductase complexed with the potent inhibitor zenarestat.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1IEJ
 
 | | OVOTRANSFERRIN, N-TERMINAL LOBE, HOLO FORM, AT 1.65 A RESOLUTION | | Descriptor: | CARBONATE ION, FE (III) ION, OVOTRANSFERRIN | | Authors: | Mizutani, K, Mikami, B, Hirose, M. | | Deposit date: | 2001-04-10 | | Release date: | 2001-06-20 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Domain closure mechanism in transferrins: new viewpoints about the hinge structure and motion as deduced from high resolution crystal structures of ovotransferrin N-lobe.
J.Mol.Biol., 309, 2001
|
|
1IEK
 
 | | SOLUTION STRUCTURE OF THE DNA DUPLEX D(CCACCGGAAC).(GTTCCGGTGG) WITH A CHIRAL ALKYL-PHOSPHONATE MOIETY (DIAESTEREOISOMER S) | | Descriptor: | 5'-D(*CP*CP*AP*CP*CP*(OCT)GP*GP*AP*AP*C)-3', 5'-D(*GP*TP*TP*CP*CP*GP*GP*TP*GP*G)-3', N-OCTANE | | Authors: | Soliva, R, Monaco, V, Gomez-Pinto, I, Meeuwenoord, N.J, van der Marel, G.A, van Boom, J.H, Gonzalez, C, Orozco, M. | | Deposit date: | 2001-04-10 | | Release date: | 2001-07-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA duplex with a chiral alkyl phosphonate moiety.
Nucleic Acids Res., 29, 2001
|
|
1IEL
 
 | | Crystal Structure of AmpC beta-lactamase from E. coli in Complex with Ceftazidime | | Descriptor: | ACYLATED CEFTAZIDIME, PHOSPHATE ION, beta-lactamase | | Authors: | Powers, R.A, Caselli, E, Focia, P.J, Prati, F, Shoichet, B.K. | | Deposit date: | 2001-04-10 | | Release date: | 2001-08-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of ceftazidime and its transition-state analogue in complex with AmpC beta-lactamase: implications for resistance mutations and inhibitor design.
Biochemistry, 40, 2001
|
|
1IEM
 
 | | Crystal Structure of AmpC beta-lactamase from E. coli in Complex with a Boronic Acid Inhibitor (1, CefB4) | | Descriptor: | PHOSPHATE ION, PINACOL[[2-AMINO-ALPHA-(1-CARBOXY-1-METHYLETHOXYIMINO)-4-THIAZOLEACETYL]AMINO]METHANEBORONATE, beta-lactamase | | Authors: | Powers, R.A, Caselli, E, Focia, P.J, Prati, F, Shoichet, B.K. | | Deposit date: | 2001-04-10 | | Release date: | 2001-08-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of ceftazidime and its transition-state analogue in complex with AmpC beta-lactamase: implications for resistance mutations and inhibitor design.
Biochemistry, 40, 2001
|
|
1IEN
 
 | | SOLUTION STRUCTURE OF TIA | | Descriptor: | PROTEIN TIA | | Authors: | Sharpe, I.A, Gehrmann, J, Loughnan, M.L, Thomas, L, Adams, D.A, Atkins, A, Palant, E, Craik, D.J, Adams, D.J, Alewood, P.F, Lewis, R.J. | | Deposit date: | 2001-04-10 | | Release date: | 2002-04-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Two new classes of conopeptides inhibit the alpha1-adrenoceptor and noradrenaline transporter.
Nat.Neurosci., 4, 2001
|
|
1IEO
 
 | | SOLUTION STRUCTURE OF MRIB-NH2 | | Descriptor: | PROTEIN MRIB-NH2 | | Authors: | Sharpe, I.A, Gehrmann, J, Loughnan, M.L, Thomas, L, Adams, D.A, Atkins, A, Palant, E, Craik, D.J, Adams, D.J, Alewood, P.F, Lewis, R.J. | | Deposit date: | 2001-04-10 | | Release date: | 2002-04-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Two new classes of conopeptides inhibit the alpha1-adrenoceptor and noradrenaline transporter.
Nat.Neurosci., 4, 2001
|
|
