17RA
 
 | |
180D
 
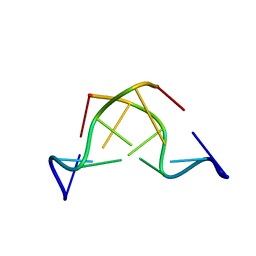 | |
180L
 
 | |
181D
 
 | |
181L
 
 | |
182D
 
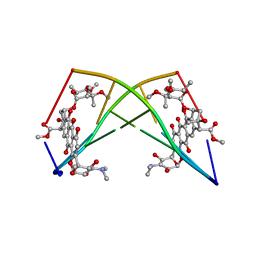 | | DNA-NOGALAMYCIN INTERACTIONS: THE CRYSTAL STRUCTURE OF D(TGATCA) COMPLEXED WITH NOGALAMYCIN | | Descriptor: | DNA (5'-D(*TP*GP*AP*TP*CP*A)-3'), NOGALAMYCIN | | Authors: | Smith, C.K, Davies, G.J, Dodson, E.J, Moore, M.H. | | Deposit date: | 1994-07-28 | | Release date: | 1994-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | DNA-nogalamycin interactions: the crystal structure of d(TGATCA) complexed with nogalamycin.
Biochemistry, 34, 1995
|
|
182L
 
 | |
183D
 
 | | X-RAY STRUCTURE OF A DNA DECAMER CONTAINING 7, 8-DIHYDRO-8-OXOGUANINE | | Descriptor: | DNA (5'-D(*CP*CP*AP*(8OG)P*CP*GP*CP*TP*GP*G)-3') | | Authors: | Lipscomb, L.A, Peek, M.E, Morningstar, M.L, Verghis, S.M, Miller, E.M, Rich, A, Essigmann, J.M, Williams, L.D. | | Deposit date: | 1994-08-01 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray structure of a DNA decamer containing 7,8-dihydro-8-oxoguanine.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
183L
 
 | |
184D
 
 | | SELF-ASSOCIATION OF A DNA LOOP CREATES A QUADRUPLEX: CRYSTAL STRUCTURE OF D(GCATGCT) AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*GP*CP*T)-3'), MAGNESIUM ION | | Authors: | Leonard, G.A, Zhang, S, Peterson, M.R, Harrop, S.J, Helliwell, J.R, Cruse, W.B.T, Langlois D'Estaintot, B, Kennard, O, Brown, T, Hunter, W.N. | | Deposit date: | 1994-08-10 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Self-association of a DNA loop creates a quadruplex: crystal structure of d(GCATGCT) at 1.8 A resolution.
Structure, 3, 1995
|
|
184L
 
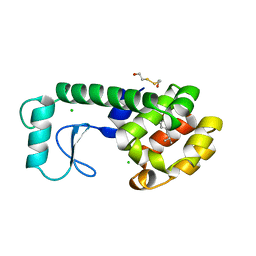 | |
185D
 
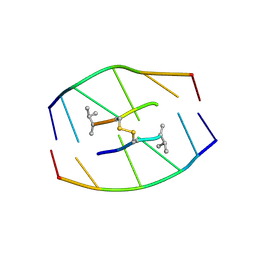 | |
185L
 
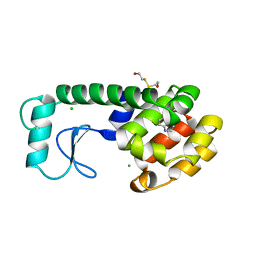 | |
186D
 
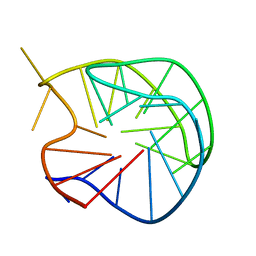 | |
186L
 
 | |
187D
 
 | |
187L
 
 | |
188D
 
 | |
188L
 
 | |
189D
 
 | |
189L
 
 | |
18GS
 
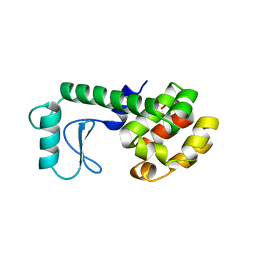
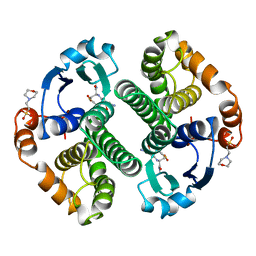 | | GLUTATHIONE S-TRANSFERASE P1-1 COMPLEXED WITH 1-(S-GLUTATHIONYL)-2,4-DINITROBENZENE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE S-(2,4 DINITROBENZENE), GLUTATHIONE S-TRANSFERASE | | Authors: | Oakley, A.J, Lo Bello, M, Ricci, G, Federici, G, Parker, M.W. | | Deposit date: | 1997-12-07 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The ligandin (non-substrate) binding site of human Pi class glutathione transferase is located in the electrophile binding site (H-site).
J.Mol.Biol., 291, 1999
|
|
190D
 
 | |
190L
 
 | |
1914
 
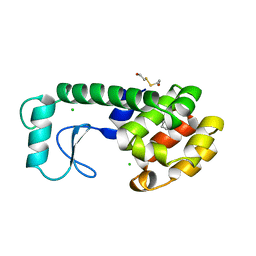
 | | SIGNAL RECOGNITION PARTICLE ALU RNA BINDING HETERODIMER, SRP9/14 | | Descriptor: | BETA-MERCAPTOETHANOL, PHOSPHATE ION, SIGNAL RECOGNITION PARTICLE 9/14 FUSION PROTEIN | | Authors: | Birse, D, Kapp, U, Strub, K, Cusack, S, Aberg, A. | | Deposit date: | 1997-11-13 | | Release date: | 1998-12-30 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | The crystal structure of the signal recognition particle Alu RNA binding heterodimer, SRP9/14.
EMBO J., 16, 1997
|
|
