1W5U
 
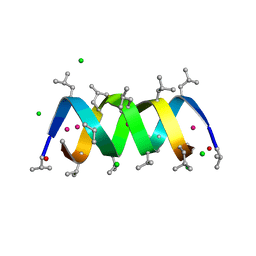 | | GRAMICIDIN D FROM BACILLUS BREVIS (ETHANOL SOLVATE) | | Descriptor: | CHLORIDE ION, ETHANOL, GRAMICIDIN D, ... | | Authors: | Glowka, M.L, Olczak, A, Bojarska, J, Szczesio, M, Duax, W.L, Burkhart, B.M, Pangborn, W.A, Langs, D.A, Wawrzak, Z. | | Deposit date: | 2004-08-10 | | Release date: | 2005-09-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Structure of Gramicidin D-Rbcl Complex at Atomic Resolution from Low-Temperature Synchrotron Data: Interactions of Double-Stranded Gramicidin Channel Contents and Cations with Channel Wall
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5FJJ
 
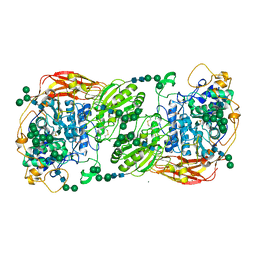 | | Three-dimensional structures of two heavily N-glycosylated Aspergillus sp. Family GH3 beta-D-glucosidases | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-GLUCOSIDASE, ... | | Authors: | Agirre, J, Ariza, A, Offen, W.A, Turkenburg, J.P, Roberts, S.M, McNicholas, S, Harris, P.V, McBrayer, B, Dohnalek, J, Cowtan, K.D, Davies, G.J, Wilson, K.S. | | Deposit date: | 2015-10-09 | | Release date: | 2016-02-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three-Dimensional Structures of Two Heavily N-Glycosylated Aspergillus Sp. Family Gh3 Beta-D-Glucosidases
Acta Crystallogr.,Sect.D, 72, 2016
|
|
5FJI
 
 | | Three-dimensional structures of two heavily N-glycosylated Aspergillus sp. Family GH3 beta-D-glucosidases | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-GLUCOSIDASE, ... | | Authors: | Agirre, J, Ariza, A, Offen, W.A, Turkenburg, J.P, Roberts, S.M, McNicholas, S, Harris, P.V, McBrayer, B, Dohnalek, J, Cowtan, K.D, Davies, G.J, Wilson, K.S. | | Deposit date: | 2015-10-09 | | Release date: | 2016-02-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three-Dimensional Structures of Two Heavily N-Glycosylated Aspergillus Sp. Family Gh3 Beta-D-Glucosidases
Acta Crystallogr.,Sect.D, 72, 2016
|
|
4Q6N
 
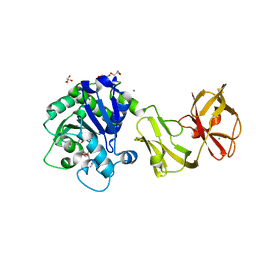 | | Structural analysis of the tripeptide-bound form of Helicobacter pylori Csd4, a D,L-carboxypeptidase | | Descriptor: | CALCIUM ION, Conserved hypothetical secreted protein, GLYCEROL, ... | | Authors: | Kim, H.S, Kim, J, Im, H.N, An, D.R, Lee, M, Hesek, D, Mobashery, S, Kim, J.Y, Cho, K, Yoon, H.J, Han, B.W, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-04-23 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a D,L-carboxypeptidase controlling the helical cell shape
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4AWE
 
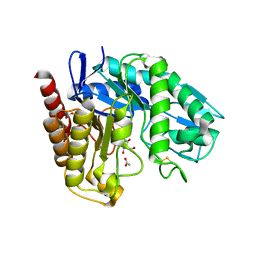 | | The Crystal Structure of Chrysonilia sitophila endo-beta-D-1,4- mannanase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goncalves, A.M.D, Silva, C.S, De Sanctis, D, Bento, I. | | Deposit date: | 2012-06-01 | | Release date: | 2012-10-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Endo-Beta-D-1,4-Mannanase from Chrysonilia Sitophila Displays a Novel Loop Arrangement for Substrate Selectivity
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1K0U
 
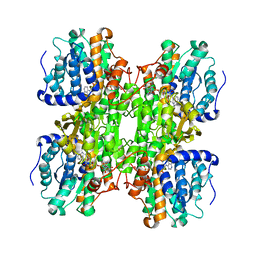 | | Inhibition of S-adenosylhomocysteine Hydrolase by "acyclic sugar" Adenosine Analogue D-eritadenine | | Descriptor: | D-ERITADENINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, S-ADENOSYL-L-HOMOCYSTEINE HYDROLASE | | Authors: | Takusagawa, F, Huang, Y, Komoto, J, Takata, Y, Gomi, T, Ogawa, H, Fujioka, M, Powell, D. | | Deposit date: | 2001-09-20 | | Release date: | 2001-10-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Inhibition of S-adenosylhomocysteine hydrolase by acyclic sugar adenosine analogue D-eritadenine. Crystal structure of S-adenosylhomocysteine hydrolase complexed with D-eritadenine.
J.Biol.Chem., 277, 2002
|
|
243D
 
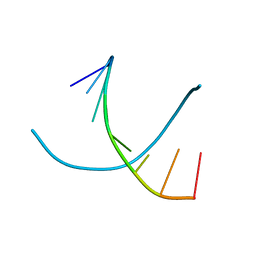 | | STRUCTURE OF THE DNA OCTANUCLEOTIDE D(ACGTACGT)2 | | Descriptor: | DNA (5'-D(*AP*CP*GP*TP*AP*CP*GP*T)-3') | | Authors: | Wilcock, D.J, Adams, A, Cardin, C.J, Wakelin, L.P.G. | | Deposit date: | 1996-01-10 | | Release date: | 1996-02-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the DNA octanucleotide d(ACGTACGT)2.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
2HK1
 
 | | Crystal structure of D-psicose 3-epimerase (DPEase) in the presence of D-fructose | | Descriptor: | D-PSICOSE 3-EPIMERASE, D-fructose, MANGANESE (II) ION | | Authors: | Kim, K, Kim, H.J, Oh, D.K, Cha, S.S, Rhee, S. | | Deposit date: | 2006-07-03 | | Release date: | 2006-08-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of d-Psicose 3-epimerase from Agrobacterium tumefaciens and its Complex with True Substrate d-Fructose: A Pivotal Role of Metal in Catalysis, an Active Site for the Non-phosphorylated Substrate, and its Conformational Changes
J.Mol.Biol., 361, 2006
|
|
4DVO
 
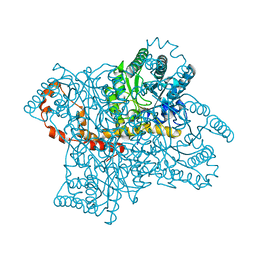 | | Room-temperature joint X-ray/neutron structure of D-xylose isomerase in complex with 2Ni2+ and per-deuterated D-sorbitol at pH 5.9 | | Descriptor: | NICKEL (II) ION, Xylose isomerase, sorbitol | | Authors: | Kovalevsky, A.Y, Hanson, L, Langan, P. | | Deposit date: | 2012-02-23 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | Inhibition of D-xylose isomerase by polyols: atomic details by joint X-ray/neutron crystallography.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4XIA
 
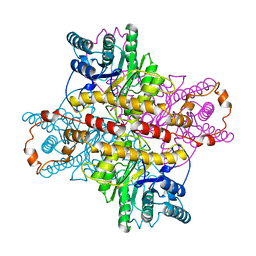 | | STRUCTURES OF D-XYLOSE ISOMERASE FROM ARTHROBACTER STRAIN B3728 CONTAINING THE INHIBITORS XYLITOL AND D-SORBITOL AT 2.5 ANGSTROMS AND 2.3 ANGSTROMS RESOLUTION, RESPECTIVELY | | Descriptor: | D-XYLOSE ISOMERASE, MAGNESIUM ION, sorbitol | | Authors: | Henrick, K, Collyer, C.A, Blow, D.M. | | Deposit date: | 1989-07-05 | | Release date: | 1990-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of D-xylose isomerase from Arthrobacter strain B3728 containing the inhibitors xylitol and D-sorbitol at 2.5 A and 2.3 A resolution, respectively.
J.Mol.Biol., 208, 1989
|
|
4RW3
 
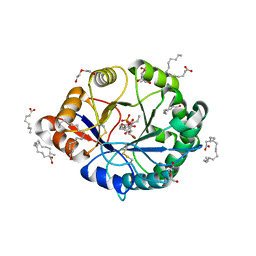 | | Structural insights into substrate binding of brown spider venom class II phospholipases D | | Descriptor: | D-MYO-INOSITOL-1-PHOSPHATE, DECANOIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Coronado, M.A, Ullah, A, da Silva, L.S, Chaves-Moreira, D, Vuitika, L, Chaim, O.M, Veiga, S.S, Chahine, J, Murakami, M.T, Arni, R.K. | | Deposit date: | 2014-12-01 | | Release date: | 2015-06-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Insights into Substrate Binding of Brown Spider Venom Class II Phospholipases D.
Curr Protein Pept Sci, 16, 2015
|
|
5XIA
 
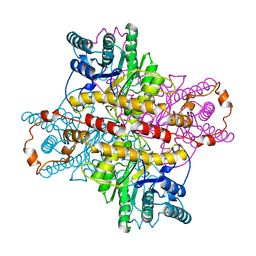 | | STRUCTURES OF D-XYLOSE ISOMERASE FROM ARTHROBACTER STRAIN B3728 CONTAINING THE INHIBITORS XYLITOL AND D-SORBITOL AT 2.5 ANGSTROMS AND 2.3 ANGSTROMS RESOLUTION, RESPECTIVELY | | Descriptor: | D-XYLOSE ISOMERASE, MAGNESIUM ION, Xylitol | | Authors: | Henrick, K, Collyer, C.A, Blow, D.M. | | Deposit date: | 1989-07-05 | | Release date: | 1990-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of D-xylose isomerase from Arthrobacter strain B3728 containing the inhibitors xylitol and D-sorbitol at 2.5 A and 2.3 A resolution, respectively.
J.Mol.Biol., 208, 1989
|
|
4UMF
 
 | | Crystal structure of 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase from Moraxella catarrhalis in complex with Magnesium ion, Phosphate ion and KDO molecule | | Descriptor: | 3-DEOXY-D-MANNO-OCTULOSONATE 8-PHOSPHATE PHOSPHATASE KDSC, 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, MAGNESIUM ION, ... | | Authors: | Dhindwal, S, Tomar, S, Kumar, P. | | Deposit date: | 2014-05-16 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Ligand-Bound Structures of 3-Deoxy-D-Manno-Octulosonate 8-Phosphate Phosphatase from Moraxella Catarrhalis Reveal a Water Channel Connecting to the Active Site for the Second Step of Catalysis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UME
 
 | | Crystal structure of 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase from Moraxella catarrhalis in complex with Magnesium ion and KDO molecule | | Descriptor: | 3-DEOXY-D-MANNO-OCTULOSONATE 8-PHOSPHATE PHOSPHATASE KDSC, 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, MAGNESIUM ION | | Authors: | Dhindwal, S, Tomar, S, Kumar, P. | | Deposit date: | 2014-05-16 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Ligand-Bound Structures of 3-Deoxy-D-Manno-Octulosonate 8-Phosphate Phosphatase from Moraxella Catarrhalis Reveal a Water Channel Connecting to the Active Site for the Second Step of Catalysis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6DZX
 
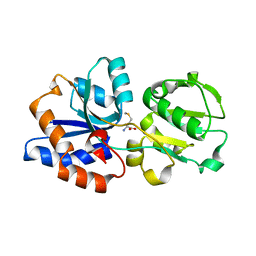 | | Crystal structure of the N. meningitides methionine-binding protein in its D-methionine bound conformation. | | Descriptor: | D-METHIONINE, Lipoprotein | | Authors: | Nguyen, P.T, Lai, J.Y, Kaiser, J.T, Rees, D.C. | | Deposit date: | 2018-07-05 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.678 Å) | | Cite: | Structures of the Neisseria meningitides methionine-binding protein MetQ in substrate-free form and bound to l- and d-methionine isomers.
Protein Sci., 28, 2019
|
|
1D67
 
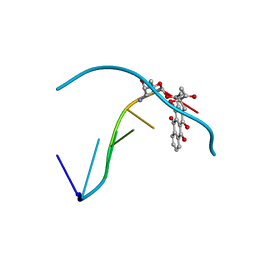 | | THE MOLECULAR STRUCTURE OF AN IDARUBICIN-D(TGATCA) COMPLEX AT HIGH RESOLUTION | | Descriptor: | DNA (5'-D(*TP*GP*AP*TP*CP*A)-3'), IDARUBICIN | | Authors: | Gallois, B, Langlois D'Estaintot, B, Brown, T, Hunter, W.N. | | Deposit date: | 1992-03-31 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of an idarubicin-d(TGATCA) complex at high resolution.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
4DUO
 
 | | Room-temperature X-ray structure of D-Xylose Isomerase in complex with 2Mg2+ ions and xylitol at pH 7.7 | | Descriptor: | MAGNESIUM ION, Xylitol, Xylose isomerase | | Authors: | Kovalevsky, A.Y, Hanson, L, Langan, P. | | Deposit date: | 2012-02-22 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of D-xylose isomerase by polyols: atomic details by joint X-ray/neutron crystallography.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4EJ0
 
 | |
4GLN
 
 | | Crystal Structure of Chemically Synthesized Heterochiral {D-Protein Antagonist plus VEGF-A} Protein Complex in space group P21/n | | Descriptor: | D-RFX001, Vascular endothelial growth factor A | | Authors: | Mandal, K, Uppalapati, M, Ault-Riche, D, Kenney, J, Lowitz, J, Sidhu, S, Kent, S.B.H. | | Deposit date: | 2012-08-14 | | Release date: | 2012-09-05 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Chemical synthesis and X-ray structure of a heterochiral {D-protein antagonist plus vascular endothelial growth factor} protein complex by racemic crystallography.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1R3Z
 
 | | Crystal structures of d(Gm5CGm5CGCGC) and d(GCGCGm5CGm5C): Effects of methylation on alternating DNA octamers | | Descriptor: | 5'-D(*GP*(5CM)P*GP*(5CM)P*GP*CP*GP*C)-3' | | Authors: | Shi, K, Pan, B, Tippin, D, Sundaralingam, M. | | Deposit date: | 2003-10-03 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of d(Gm5)CGm5CGCGC) and d(GCGCGm5CGm5C): effects of methylation on alternating DNA octamers.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1XLJ
 
 | | MECHANISM FOR ALDOSE-KETOSE INTERCONVERSION BY D-XYLOSE ISOMERASE INVOLVING RING OPENING FOLLOWED BY A 1,2-HYDRIDE SHIFT | | Descriptor: | D-XYLOSE ISOMERASE, MANGANESE (II) ION, Xylitol | | Authors: | Collyer, C.A, Henrick, K, Blow, D.M. | | Deposit date: | 1991-10-09 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism for aldose-ketose interconversion by D-xylose isomerase involving ring opening followed by a 1,2-hydride shift.
J.Mol.Biol., 212, 1990
|
|
4GLS
 
 | | Crystal Structure of Chemically Synthesized Heterochiral {D-Protein Antagonist plus VEGF-A} Protein Complex in space group P21 | | Descriptor: | D- RFX001, D- Vascular endothelial growth factor-A, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mandal, K, Uppalapati, M, Ault-Riche, D, Kenney, J, Lowitz, J, Sidhu, S, Kent, S.B.H. | | Deposit date: | 2012-08-14 | | Release date: | 2012-09-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Chemical synthesis and X-ray structure of a heterochiral {D-protein antagonist plus vascular endothelial growth factor} protein complex by racemic crystallography.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1R41
 
 | | Crystal structures of d(Gm5CGm5CGCGC) and d(GCGCGm5CGm5C): Effects of methylation on alternating DNA octamers | | Descriptor: | 5'-D(*GP*CP*GP*CP*GP*(5CM)P*GP*(5CM))-3' | | Authors: | Shi, K, Pan, B, Tippin, D, Sundaralingam, M. | | Deposit date: | 2003-10-03 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of d(Gm5)CGm5CGCGC) and d(GCGCGm5CGm5C): effects of methylation on alternating DNA octamers.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1T0A
 
 | | Crystal Structure of 2C-Methyl-D-Erythritol-2,4-cyclodiphosphate Synthase from Shewanella Oneidensis | | Descriptor: | 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, COBALT (II) ION, FARNESYL DIPHOSPHATE, ... | | Authors: | Ni, S, Robinson, H, Marsing, G.C, Bussiere, D.E, Kennedy, M.A. | | Deposit date: | 2004-04-08 | | Release date: | 2004-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of 2C-methyl-D-erythritol-2,4-cyclodiphosphate synthase from Shewanella oneidensis at 1.6 A: identification of farnesyl pyrophosphate trapped in a hydrophobic cavity.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1XLF
 
 | | MECHANISM FOR ALDOSE-KETOSE INTERCONVERSION BY D-XYLOSE ISOMERASE INVOLVING RING OPENING FOLLOWED BY A 1,2-HYDRIDE SHIFT | | Descriptor: | D-XYLOSE ISOMERASE, D-gluconic acid, MANGANESE (II) ION | | Authors: | Collyer, C.A, Henrick, K, Blow, D.M. | | Deposit date: | 1991-10-09 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism for aldose-ketose interconversion by D-xylose isomerase involving ring opening followed by a 1,2-hydride shift.
J.Mol.Biol., 212, 1990
|
|
