8KEU
 
 | |
3ML0
 
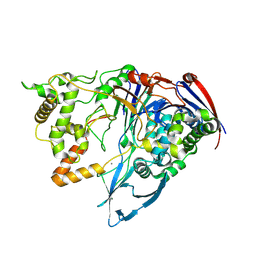 | | Thermostable Penicillin G acylase from Alcaligenes faecalis in tetragonal form | | Descriptor: | CALCIUM ION, Penicillin G acylase, alpha subunit, ... | | Authors: | Varshney, N.K, Kumar, R.S, Ignatova, Z, Dodson, E, Suresh, C.G. | | Deposit date: | 2010-04-16 | | Release date: | 2010-05-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystallization and X-ray structure analysis of a thermostable penicillin G acylase from Alcaligenes faecalis.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3MPJ
 
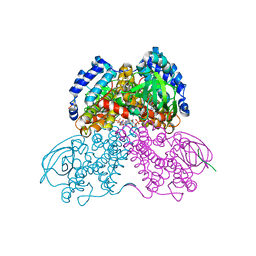 | | Structure of the glutaryl-coenzyme A dehydrogenase | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Glutaryl-CoA dehydrogenase, ... | | Authors: | Wischgoll, S, Warkentin, E, Boll, M, Ermler, U. | | Deposit date: | 2010-04-27 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for promoting and preventing decarboxylation in glutaryl-coenzyme a dehydrogenases.
Biochemistry, 49, 2010
|
|
4LZT
 
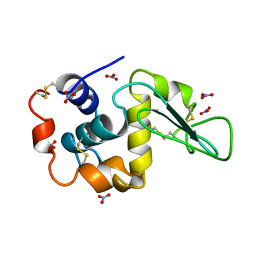 | | ATOMIC RESOLUTION REFINEMENT OF TRICLINIC HEW LYSOZYME AT 295K | | Descriptor: | LYSOZYME, NITRATE ION | | Authors: | Walsh, M.A, Schneider, T, Sieker, L.C, Dauter, Z, Lamzin, V, Wilson, K.S. | | Deposit date: | 1997-03-31 | | Release date: | 1998-04-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Refinement of triclinic hen egg-white lysozyme at atomic resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
4LXJ
 
 | | Saccharomyces cerevisiae lanosterol 14-alpha demethylase with lanosterol bound | | Descriptor: | LANOSTEROL, Lanosterol 14-alpha demethylase, OXYGEN MOLECULE, ... | | Authors: | Monk, B.C, Tomasiak, T.M, Keniya, M.V, Huschmann, F.U, Tyndall, J.D.A, O'Connell III, J.D, Cannon, R.D, McDonald, J, Rodriguez, A, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 2013-07-29 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Architecture of a single membrane spanning cytochrome P450 suggests constraints that orient the catalytic domain relative to a bilayer.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4M6D
 
 | | Crystal structure of the aptamer minF-lysozyme complex. | | Descriptor: | Lysozyme C, aptamer | | Authors: | Malashkevich, V.N, Padlan, F.C, Toro, R, Girvin, M, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-09 | | Release date: | 2013-12-11 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal structure of the aptamer minF-lysozyme complex.
To be Published
|
|
3M6D
 
 | | The crystal structure of the d307a mutant of glycoside Hydrolase (family 31) from ruminococcus obeum atcc 29174 | | Descriptor: | Uncharacterized protein | | Authors: | Tan, K, Tesar, C, Freeman, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-15 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Novel alpha-glucosidase from human gut microbiome: substrate specificities and their switch.
Faseb J., 24, 2010
|
|
3M83
 
 | |
4MB4
 
 | | Crystal structure of E153Q mutant of cold-adapted chitinase from Moritella complex with Nag4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase 60, GLYCEROL, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Rypniewski, W. | | Deposit date: | 2013-08-19 | | Release date: | 2014-03-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.481 Å) | | Cite: | Crystal structures of substrate-bound chitinase from the psychrophilic bacterium Moritella marina and its structure in solution
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4M82
 
 | | The structure of E292S glycosynthase variant of exo-1,3-beta-glucanase from Candida albicans complexed with p-nitrophenyl-gentiobioside (product) at 1.6A resolution | | Descriptor: | 1,2-ETHANEDIOL, 4-nitrophenyl 6-O-beta-D-glucopyranosyl-beta-D-glucopyranoside, EXO-1,3-BETA-GLUCANASE, ... | | Authors: | Nakatani, Y, Cutfield, S.M, Larsen, D.S, Cutfield, J.F. | | Deposit date: | 2013-08-12 | | Release date: | 2014-06-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.592 Å) | | Cite: | Major Change in Regiospecificity for the Exo-1,3-beta-glucanase from Candida albicans following Its Conversion to a Glycosynthase.
Biochemistry, 53, 2014
|
|
3MDW
 
 | | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-formimino-L-Aspartate | | Descriptor: | GLYCEROL, N-[(E)-iminomethyl]-L-aspartic acid, N-formimino-L-Glutamate Iminohydrolase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Marti-Arbona, R, Raushel, F.M, Almo, S.C. | | Deposit date: | 2010-03-30 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8979 Å) | | Cite: | Structure of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa.
Biochemistry, 54, 2015
|
|
3MEQ
 
 | | Crystal structure of alcohol dehydrogenase from Brucella melitensis | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Alcohol dehydrogenase, ... | | Authors: | Arakaki, T.L, Staker, B.L, Gardberg, A, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of alcohol dehydrogenase from Brucella melitensis
To be Published
|
|
4MB5
 
 | | Crystal structure of E153Q mutant of cold-adapted chitinase from Moritella complex with Nag5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Chitinase 60, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Rypniewski, W. | | Deposit date: | 2013-08-19 | | Release date: | 2014-03-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.639 Å) | | Cite: | Crystal structures of substrate-bound chitinase from the psychrophilic bacterium Moritella marina and its structure in solution
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3M18
 
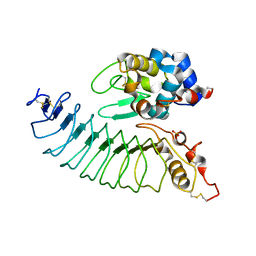 | |
4MCA
 
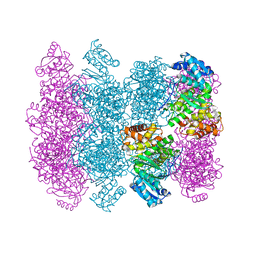 | |
3MMS
 
 | | Crystal structure of Streptococcus pneumoniae MTA/SAH nucleosidase in complex with 8-aminoadenine | | Descriptor: | 5'-methylthioadenosine / S-adenosylhomocysteine nucleosidase, 9H-purine-6,8-diamine, GLYCEROL | | Authors: | Siu, K.K.W, Lee, J.E, Horvatin-Mrakovcic, C, Howell, P.L. | | Deposit date: | 2010-04-20 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Streptococcus pneumoniae MTA/SAH nucleosidase in complex with 8-aminoadenine
TO BE PUBLISHED
|
|
3MN1
 
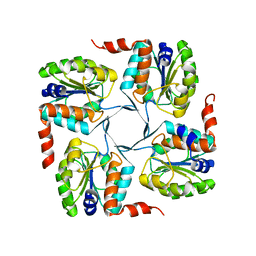 | | Crystal structure of probable yrbi family phosphatase from pseudomonas syringae pv.phaseolica 1448a | | Descriptor: | CHLORIDE ION, probable yrbi family phosphatase | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Freeman, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-20 | | Release date: | 2010-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the divergence of substrate specificity and biological function within HAD phosphatases in lipopolysaccharide and sialic acid biosynthesis.
Biochemistry, 52, 2013
|
|
3M4V
 
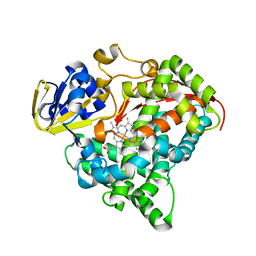 | | Crystal structure of the A330P mutant of cytochrome P450 BM3 | | Descriptor: | Bifunctional P-450/NADPH-P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yang, W, Whitehouse, C.J.C, Bell, S.G, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-03-12 | | Release date: | 2011-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the properties of two single-site proline mutants of CYP102A1 (P450BM3)
Chembiochem, 11, 2010
|
|
4J6B
 
 | |
3MSD
 
 | | Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. | | Descriptor: | ACETATE ION, GLYCEROL, Intra-cellular xylanase ixt6, ... | | Authors: | Solomon, V, Zolotnitsky, G, Alhadeff, R, Shoham, Y, Shoham, G. | | Deposit date: | 2010-04-29 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus.
TO BE PUBLISHED
|
|
4JFU
 
 | | Crystal structure of a bacterial fucosidase with iminosugar inhibitor | | Descriptor: | (2S,3R,4S,5S)-2-methyl-5-(4-methylphenyl)pyrrolidine-3,4-diol, GLYCEROL, IMIDAZOLE, ... | | Authors: | Wright, D.W, Davies, G.J. | | Deposit date: | 2013-02-28 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | alpha-L-fucosidase inhibition by pyrrolidine-ferrocene hybrids: rationalization of ligand-binding properties by structural studies.
Chemistry, 19, 2013
|
|
4JBH
 
 | | 2.2A resolution structure of cobalt and zinc bound thermostable alcohol dehydrogenase from Pyrobaculum aerophilum | | Descriptor: | Alcohol dehydrogenase (Zinc), CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Lovell, S, Battaile, K.P, Vitale, A, Throne, N, Hu, X, Shen, M, D'Auria, S, Auld, D.S. | | Deposit date: | 2013-02-19 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Physicochemical Characterization of a Thermostable Alcohol Dehydrogenase from Pyrobaculum aerophilum.
Plos One, 8, 2013
|
|
3M9S
 
 | | Crystal structure of respiratory complex I from Thermus thermophilus | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Efremov, R.G, Baradaran, R, Sazanov, L.A. | | Deposit date: | 2010-03-22 | | Release date: | 2010-05-26 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | The architecture of respiratory complex I
Nature, 465, 2010
|
|
4JKK
 
 | |
4JQA
 
 | | AKR1C2 complex with mefenamic acid | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2,3-DIMETHYLPHENYL)AMINO]BENZOIC ACID, Aldo-keto reductase family 1 member C2, ... | | Authors: | Yosaatmadja, Y, Flanagan, J.U, Squire, C.J. | | Deposit date: | 2013-03-20 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural basis of NSAID selectivity for the aldo-keto reductase 1C family
To be Published
|
|
