3A3V
 
 | | Crystal structure of reducing-end-xylose releasing exo-oligoxylanase Y198F mutant | | Descriptor: | GLYCEROL, NICKEL (II) ION, Xylanase Y | | Authors: | Hidaka, M, Fushinobu, S, Honda, Y, Kitaoka, M. | | Deposit date: | 2009-06-22 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural explanation for the acquisition of glycosynthase activity
J.Biochem., 147, 2010
|
|
1P3Y
 
 | | MrsD from Bacillus sp. HIL-Y85/54728 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MrsD protein | | Authors: | Blaesse, M, Kupke, T, Huber, R, Steinbacher, S. | | Deposit date: | 2003-04-19 | | Release date: | 2003-08-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structure of MrsD, an FAD-binding protein of the HFCD family.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
2WOW
 
 | | Trypanosoma brucei trypanothione reductase with NADP and trypanothione bound | | Descriptor: | BIS(GAMMA-GLUTAMYL-CYSTEINYL-GLYCINYL)SPERMIDINE, FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Alphey, M.S, Fairlamb, A.H. | | Deposit date: | 2009-07-30 | | Release date: | 2010-08-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dihydroquinazolines as a Novel Class of Trypanosoma Brucei Trypanothione Reductase Inhibitors: Discovery, Synthesis, and Characterization of Their Binding Mode by Protein Crystallography.
J.Med.Chem., 54, 2011
|
|
2WP5
 
 | | Trypanosoma brucei trypanothione reductase in complex with 3,4- dihydroquinazoline inhibitor (DDD00065414) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Alphey, M.S, Patterson, S, Fairlamb, A.H. | | Deposit date: | 2009-08-03 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dihydroquinazolines as a Novel Class of Trypanosoma Brucei Trypanothione Reductase Inhibitors: Discovery, Synthesis, and Characterization of Their Binding Mode by Protein Crystallography.
J.Med.Chem., 54, 2011
|
|
3EVX
 
 | | Crystal structure of the human E2-like ubiquitin-fold modifier conjugating enzyme 1 (Ufc1). Northeast Structural Genomics Consortium target HR41 | | Descriptor: | THIOCYANATE ION, Ufm1-conjugating enzyme 1 | | Authors: | Forouhar, F, Abashidze, M, Seetharaman, J, Ho, C.K, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-10-13 | | Release date: | 2008-10-21 | | Last modified: | 2023-01-04 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | NMR and X-RAY structures of human E2-like ubiquitin-fold modifier conjugating enzyme 1 (UFC1) reveal structural and functional conservation in the metazoan UFM1-UBA5-UFC1 ubiquination pathway.
J.STRUCT.FUNCT.GENOM., 10, 2009
|
|
1OQ0
 
 | |
1ORY
 
 | | FLAGELLAR EXPORT CHAPERONE IN COMPLEX WITH ITS COGNATE BINDING PARTNER | | Descriptor: | Flagellin, PHOSPHATE ION, flagellar protein FliS | | Authors: | Evdokimov, A.G, Phan, J, Tropea, J.E, Routzahn, K.M, Peters III, H.K, Pokross, M, Waugh, D.S. | | Deposit date: | 2003-03-17 | | Release date: | 2003-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Similar modes of polypeptide recognition by export chaperones in flagellar biosynthesis and type III secretion
Nat.Struct.Biol., 10, 2003
|
|
1OTH
 
 | | CRYSTAL STRUCTURE OF HUMAN ORNITHINE TRANSCARBAMOYLASE COMPLEXED WITH N-PHOSPHONACETYL-L-ORNITHINE | | Descriptor: | N-(PHOSPHONOACETYL)-L-ORNITHINE, PROTEIN (ORNITHINE TRANSCARBAMOYLASE) | | Authors: | Shi, D, Morizono, H, Ha, Y, Aoyagi, M, Tuchman, N, Allewell, N.M. | | Deposit date: | 1998-10-06 | | Release date: | 1999-10-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85-A resolution crystal structure of human ornithine transcarbamoylase complexed with N-phosphonacetyl-L-ornithine. Catalytic mechanism and correlation with inherited deficiency.
J.Biol.Chem., 273, 1998
|
|
3A9L
 
 | | Structure of Bacteriophage poly-gamma-glutamate hydrolase | | Descriptor: | PHOSPHATE ION, Poly-gamma-glutamate hydrolase, ZINC ION | | Authors: | Fujimoto, Z, Kimura, K. | | Deposit date: | 2009-10-30 | | Release date: | 2010-11-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of bacteriophage PhiNIT1 zinc peptidase PghP that hydrolyzes gamma-glutamyl linkage of bacterial poly-gamma-glutamate
Proteins, 80, 2012
|
|
2WZB
 
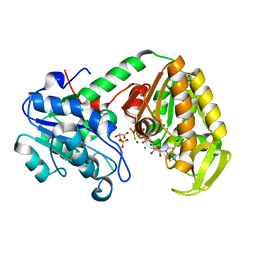 | | The catalytically active fully closed conformation of human phosphoglycerate kinase in complex with ADP, 3PG and magnesium trifluoride | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Bowler, M.W, Cliff, M.J, Marston, J.P.M, Baxter, N.J, Hownslow, A.M.H, Varga, A.V, Szabo, J, Vas, M, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-11-27 | | Release date: | 2010-04-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Transition State Analogue Structures of Human Phosphoglycerate Kinase Establish the Importance of Charge Balance in Catalysis.
J.Am.Chem.Soc., 132, 2010
|
|
3P9W
 
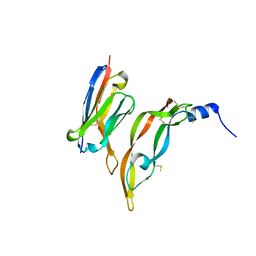 | |
1LQG
 
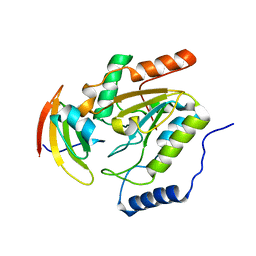 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE COMPLEX WITH URACIL-DNA GLYCOSYLASE INHIBITOR PROTEIN | | Descriptor: | URACIL-DNA GLYCOSYLASE, URACIL-DNA GLYCOSYLASE INHIBITOR | | Authors: | Saikrishnan, K, Sagar, M.B, Ravishankar, R, Roy, S, Purnapatre, K, Handa, P, Varshney, U, Vijayan, M. | | Deposit date: | 2002-05-10 | | Release date: | 2002-11-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Domain closure and action of uracil DNA glycosylase (UDG): structures of new crystal forms containing the Escherichia coli enzyme and a comparative study of the known structures involving UDG.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
2W8X
 
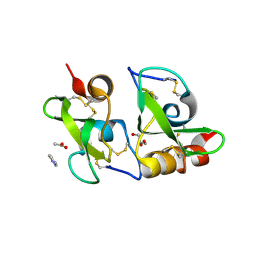 | | Structure of the tick ion-channel modulator Ra-KLP | | Descriptor: | ACETATE ION, GLYCEROL, IMIDAZOLE, ... | | Authors: | Paesen, G.C, Siebold, C, Dallas, M, Peers, C, Harlos, K, Nuttall, P.A, Nunn, M.A, Stuart, D.I, Esnouf, R.M. | | Deposit date: | 2009-01-20 | | Release date: | 2009-05-05 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An Ion-Channel Modulator from the Saliva of the Brown Ear Tick Has a Highly Modified Kunitz/Bpti Structure.
J.Mol.Biol., 389, 2009
|
|
1UIB
 
 | | ANALYSIS OF THE STABILIZATION OF HEN LYSOZYME WITH THE HELIX DIPOLE AND CHARGED SIDE CHAINS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LYSOZYME | | Authors: | Motoshima, H, Ohmura, T, Ueda, T, Imoto, T. | | Deposit date: | 1996-11-26 | | Release date: | 1997-11-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Fluctuations in free or substrate-complexed lysozyme and a mutant of it detected on x-ray crystallography and comparison with those detected on NMR.
J.Biochem.(Tokyo), 131, 2002
|
|
1UIH
 
 | | ANALYSIS OF THE STABILIZATION OF HEN LYSOZYME WITH THE HELIX DIPOLE AND CHARGED SIDE CHAINS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LYSOZYME | | Authors: | Motoshima, H, Ohmura, T, Ueda, T, Imoto, T. | | Deposit date: | 1996-11-26 | | Release date: | 1997-11-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Fluctuations in free or substrate-complexed lysozyme and a mutant of it detected on x-ray crystallography and comparison with those detected on NMR.
J.Biochem.(Tokyo), 131, 2002
|
|
3FE3
 
 | |
2VTV
 
 | | PhaZ7 depolymerase from Paucimonas lemoignei | | Descriptor: | GLYCEROL, PHB depolymerase PhaZ7 | | Authors: | Papageorgiou, A.C, Hermawan, S, Singh, C.B, Jendrossek, D. | | Deposit date: | 2008-05-16 | | Release date: | 2008-08-26 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of poly(3-hydroxybutyrate) hydrolysis by PhaZ7 depolymerase from Paucimonas lemoignei.
J. Mol. Biol., 382, 2008
|
|
1OK4
 
 | | Archaeal fructose 1,6-bisphosphate aldolase covalently bound to the substrate dihydroxyacetone phosphate | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, FRUCTOSE-BISPHOSPHATE ALDOLASE CLASS I | | Authors: | Lorentzen, E, Zwart, P, Stark, A, Hensel, R, Siebers, B, Pohl, E. | | Deposit date: | 2003-07-17 | | Release date: | 2003-09-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an archaeal class I aldolase and the evolution of (betaalpha)8 barrel proteins.
J. Biol. Chem., 278, 2003
|
|
1OK6
 
 | | Orthorhombic crystal form of an Archaeal fructose 1,6-bisphosphate aldolase | | Descriptor: | FRUCTOSE-BISPHOSPHATE ALDOLASE CLASS I, GLYCEROL | | Authors: | Lorentzen, E, Zwart, P, Stark, A, Hensel, R, Siebers, B, Pohl, E. | | Deposit date: | 2003-07-18 | | Release date: | 2003-09-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of an archaeal class I aldolase and the evolution of (betaalpha)8 barrel proteins.
J. Biol. Chem., 278, 2003
|
|
2K0S
 
 | |
1OL0
 
 | | Crystal structure of a camelised human VH | | Descriptor: | GLYCEROL, IMMUNOGLOBULIN G, SULFATE ION | | Authors: | Dottorini, T, Vaughan, C.K, Walsh, M.A, Losurdo, P, Sollazzo, M. | | Deposit date: | 2003-08-02 | | Release date: | 2004-01-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of a Human Vh: Requirements for Maintaining a Monomeric Fragment
Biochemistry, 43, 2004
|
|
2ICX
 
 | | Crystal Structure of a Putative UDP-glucose Pyrophosphorylase from Arabidopsis Thaliana with Bound UTP | | Descriptor: | DIMETHYL SULFOXIDE, Probable UTP-glucose-1-phosphate uridylyltransferase 2, URIDINE 5'-TRIPHOSPHATE | | Authors: | McCoy, J.G, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-09-13 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Dynamics of UDP-Glucose Pyrophosphorylase from Arabidopsis thaliana with Bound UDP-Glucose and UTP.
J.Mol.Biol., 366, 2007
|
|
1UN1
 
 | | Xyloglucan endotransglycosylase native structure. | | Descriptor: | GOLD ION, XYLOGLUCAN ENDOTRANSGLYCOSYLASE, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Johansson, P, Brumer, H, Kallas, A, Henriksson, H, Denman, S, Teeri, T.T, Jones, T.A. | | Deposit date: | 2003-09-03 | | Release date: | 2004-03-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of a Poplar Xyloglucan Endotransglycosylase Reveal Details of Transglycosylation Acceptor Binding
Plant Cell, 16, 2004
|
|
1ONR
 
 | | STRUCTURE OF TRANSALDOLASE B | | Descriptor: | TRANSALDOLASE B | | Authors: | Jia, J, Huang, W, Lindqvist, Y, Schneider, G. | | Deposit date: | 1996-08-13 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of transaldolase B from Escherichia coli suggests a circular permutation of the alpha/beta barrel within the class I aldolase family.
Structure, 4, 1996
|
|
2W10
 
 | | Mona SH3C in complex | | Descriptor: | GRB2-RELATED ADAPTOR PROTEIN 2, PHOSPHATE ION, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 23 | | Authors: | Harkiolaki, M, Feller, S.M. | | Deposit date: | 2008-10-13 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Distinct Binding Modes of Two Epitopes in Gab2 that Interact with the Sh3C Domain of Grb2.
Structure, 17, 2009
|
|
