9GQ3
 
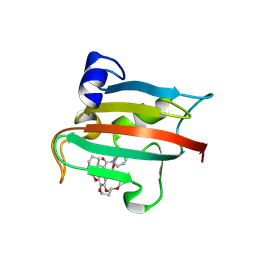 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog p5(1,4)-(E) | | Descriptor: | (2~{S},9~{S},13~{E})-2-cyclohexyl-22,25-dimethoxy-11,16,20-trioxa-4-azatricyclo[19.2.2.0^{4,9}]pentacosa-1(24),13,21(25),22-tetraene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Spiske, M, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformationally Restricted Macrocycles as Improved FKBP51 Inhibitors Enabled by Systematic Linker Derivatization.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
6T5P
 
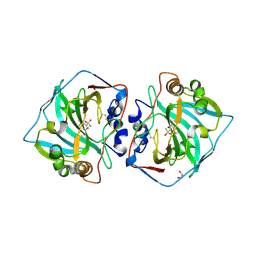 | | Human Carbonic anhydrase XII bound by 3,5-Di-tert-butylbenzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 3,5-di~{tert}-butylbenzenesulfonamide, Carbonic anhydrase 12, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2019-10-16 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Isoform-Selective Enzyme Inhibitors by Exploring Pocket Size According to the Lock-and-Key Principle.
Biophys.J., 119, 2020
|
|
6X9I
 
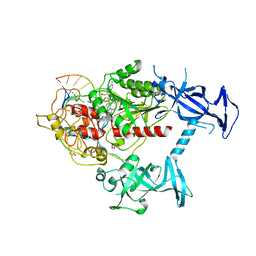 | | Human DNMT1(729-1600) Bound to Zebularine-Containing 12mer dsDNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*AP*GP*GP*CP*(5CM)P*GP*CP*CP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*GP*G)-R(P*(PYO))-D(P*GP*GP*CP*CP*TP*C)-3'), ... | | Authors: | Pathuri, S, Horton, J.R, Cheng, X. | | Deposit date: | 2020-06-02 | | Release date: | 2021-07-07 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a first-in-class reversible DNMT1-selective inhibitor with improved tolerability and efficacy in acute myeloid leukemia.
Nat Cancer, 2, 2021
|
|
4XEB
 
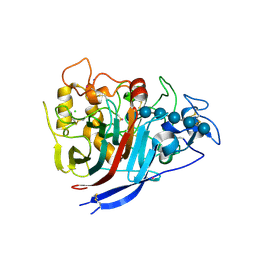 | | The structure of P. funicolosum Cel7A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Glucanase, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2014-12-23 | | Release date: | 2016-06-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Engineering enhanced cellobiohydrolase activity
Nat Commun, 9(1), 2018
|
|
9GQ6
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog p5(1,2)-H2 | | Descriptor: | (2~{S},9~{S})-2-cyclohexyl-19,22-dimethoxy-11,17-dioxa-4-azatricyclo[16.2.2.0^{4,9}]docosa-1(20),18,21-triene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Spiske, M, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Conformationally Restricted Macrocycles as Improved FKBP51 Inhibitors Enabled by Systematic Linker Derivatization.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
5HLK
 
 | | Crystal structure of the ternary EcoRV-DNA-Lu complex with cleaved DNA substrate. | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*AP*AP*GP*AP*TP)-3'), DNA (5'-D(*AP*TP*CP*TP*TP*TP)-3'), ... | | Authors: | Sangani, S.S, Kehr, A.D, Sinha, K, Rule, G.S, Jen-Jacobson, L. | | Deposit date: | 2016-01-15 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metal Ion Binding at the Catalytic Site Induces Widely Distributed Changes in a Sequence Specific Protein-DNA Complex.
Biochemistry, 55, 2016
|
|
5KWH
 
 | | Crystal structure of CK2 | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase II subunit alpha, SULFATE ION, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2016-07-18 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structure of CK2
To Be Published
|
|
9GQ7
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog p5(2,1)-(E) | | Descriptor: | (2~{S},9~{S},14~{E})-2-cyclohexyl-19,22-dimethoxy-11,17-dioxa-4-azatricyclo[16.2.2.0^{4,9}]docosa-1(20),14,18,21-tetraene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Spiske, M, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformationally Restricted Macrocycles as Improved FKBP51 Inhibitors Enabled by Systematic Linker Derivatization.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
6TDG
 
 | | Crystal structure of Aspergillus fumigatus Glucosamine-6-phosphate N-acetyltransferase 1 in complex with compound 2 | | Descriptor: | 2-chloranyl-3-(4~{H}-1,2,4-triazol-3-yl)aniline, ACETYL COENZYME *A, Glucosamine 6-phosphate N-acetyltransferase | | Authors: | Raimi, O.G, Stanley, M, Lockhart, D. | | Deposit date: | 2019-11-08 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Targeting a critical step in fungal hexosamine biosynthesis.
J.Biol.Chem., 295, 2020
|
|
9GQ4
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog p5(3,1)-(E) | | Descriptor: | (2~{S},9~{S},16~{E})-2-cyclohexyl-21,24-dimethoxy-11,14,19-trioxa-4-azatricyclo[18.2.2.0^{4,9}]tetracosa-1(23),16,20(24),21-tetraene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Spiske, M, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformationally Restricted Macrocycles as Improved FKBP51 Inhibitors Enabled by Systematic Linker Derivatization.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9GPY
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog 12g/j | | Descriptor: | 1,2-ETHANEDIOL, 2-cyclohexyl-18,19-dimethoxy-13-methyl-11,17-dioxa-4-aza-1lambda5,18lambda5-diphospha-1,15-distannatetracyclo[13.4.0.04,9.016,20]nonadecane-3,10-quinone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Buffa, V, Walz, C, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conformational Plasticity and Binding Affinity Enhancement Controlled by Linker Derivatization in Macrocycles.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9GQB
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog p5(2,1)-Diol-II | | Descriptor: | (2~{S},9~{S},14~{S},15~{S})-2-cyclohexyl-19,22-dimethoxy-14,15-bis(oxidanyl)-11,17-dioxa-4-azatricyclo[16.2.2.0^{4,9}]docosa-1(21),18(22),19-triene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Spiske, M, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformationally Restricted Macrocycles as Improved FKBP51 Inhibitors Enabled by Systematic Linker Derivatization.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
8ZIK
 
 | | Crystal structure of a beta-1,4-endoglucanase from Bispora sp. MEY-1 in complex with cellotetraose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, cellulase | | Authors: | Zheng, J, Luo, H.Y, Yao, B, Tian, J. | | Deposit date: | 2024-05-14 | | Release date: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Unraveling the Key Determinants of Substrate Promiscuity in Glycoside Hydrolase Family 5_5 Cellulases
To Be Published
|
|
6GRO
 
 | | Human CSNK1G3 bound to SB-223133 | | Descriptor: | 1,2-ETHANEDIOL, 4-[5-(2-azanylpyrimidin-4-yl)-4-(4-fluorophenyl)imidazol-1-yl]cyclohexan-1-ol, CHLORIDE ION, ... | | Authors: | Szklarz, M, Vollmar, M, von Delft, F, Bountra, C, Knapp, S, Edwards, A.M, Arrowsmith, C, Elkins, J.M. | | Deposit date: | 2018-06-12 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | CSNK1G3 bound to SB-223133
To Be Published
|
|
9GQ2
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog p5(1,1)-(E) | | Descriptor: | (2~{S},9~{S},13~{E})-2-cyclohexyl-18,21-dimethoxy-11,16-dioxa-4-azatricyclo[15.2.2.0^{4,9}]henicosa-1(19),13,17,20-tetraene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Spiske, M, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Conformationally Restricted Macrocycles as Improved FKBP51 Inhibitors Enabled by Systematic Linker Derivatization.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
6HHO
 
 | | Crystal structure of RIP1 kinase in complex with GSK547 | | Descriptor: | 6-[4-[(5~{S})-5-[3,5-bis(fluoranyl)phenyl]pyrazolidin-1-yl]carbonylpiperidin-1-yl]pyrimidine-4-carbonitrile, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Thorpe, J.H, Harris, P.A. | | Deposit date: | 2018-08-28 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | RIP1 Kinase Drives Macrophage-Mediated Adaptive Immune Tolerance in Pancreatic Cancer.
Cancer Cell, 34, 2018
|
|
5KM9
 
 | |
5HNX
 
 | | Structural basis of backwards motion in kinesin-14: minus-end directed nKn664 in the nucleotide-free state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Shigematsu, H, Yokoyama, T, Kikkawa, M, Shirouzu, M, Nitta, R. | | Deposit date: | 2016-01-19 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural Basis of Backwards Motion in Kinesin-1-Kinesin-14 Chimera: Implication for Kinesin-14 Motility
Structure, 24, 2016
|
|
6WVO
 
 | | Crystal Structure of Recombinant Human Acetylcholinesterase In Complex with GD and HI-6 | | Descriptor: | (1R)-1,2,2-TRIMETHYLPROPYL (S)-METHYLPHOSPHINATE, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | McGuire, J.R, Bester, S.M, Pegan, S.D, Height, J.J. | | Deposit date: | 2020-05-06 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural and Biochemical Insights into the Inhibition of Human Acetylcholinesterase by G-Series Nerve Agents and Subsequent Reactivation by HI-6.
Chem.Res.Toxicol., 34, 2021
|
|
7NTK
 
 | | PALS1 PDZ1 domain with SARS-CoV-2_E PBM complex | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, Envelope small membrane protein, ... | | Authors: | Javorsky, A, Kvansakul, M. | | Deposit date: | 2021-03-10 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of coronavirus E protein interactions with human PALS1 PDZ domain.
Commun Biol, 4, 2021
|
|
7S26
 
 | | ROCK1 IN COMPLEX WITH LIGAND G5018 | | Descriptor: | 2-[methyl(phenyl)amino]-1-[4-(1H-pyrrolo[2,3-b]pyridin-3-yl)-3,6-dihydropyridin-1(2H)-yl]ethan-1-one, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Rho-associated protein kinase 1 | | Authors: | Ganichkin, O, Harris, S.F, Steinbacher, S. | | Deposit date: | 2021-09-03 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.744 Å) | | Cite: | Chemical space docking enables large-scale structure-based virtual screening to discover ROCK1 kinase inhibitors.
Nat Commun, 13, 2022
|
|
8TQ7
 
 | | Crystal structure of Fab.34.2.12 in complex with MHC-I (H2-Dd) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, Fab 34.2.12 Light Chain, ... | | Authors: | Jiang, J, Boyd, L.F, Natarajan, K, Margulies, D.H. | | Deposit date: | 2023-08-06 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Experimental Structures of Antibody/MHC-I Complexes Reveal Details of Epitopes Overlooked by Computational Prediction.
J Immunol., 212, 2024
|
|
9MJM
 
 | | SOS1 IN COMPLEX WITH AN INHIBITOR | | Descriptor: | 1,2-ETHANEDIOL, 2-({(1R)-1-[2-methyl-3-(trifluoromethyl)phenyl]ethyl}amino)-3-(2-oxaspiro[3.3]heptan-6-yl)-5,6,7,8-tetrahydropyrido[4,3-d]pyrimidin-4(3H)-one, IMIDAZOLE, ... | | Authors: | Bell, J.A. | | Deposit date: | 2024-12-16 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Exploiting Solvent Exposed Salt-Bridge Interactions for the Discovery of Potent Inhibitors of SOS1 Using Free-Energy Perturbation Simulations.
Acs Med.Chem.Lett., 16, 2025
|
|
9CK2
 
 | | Human PU.1 minimal ETS Domain (165-258) bound to d(AATAAGCGGAAGTGGG) | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*AP*GP*CP*GP*GP*AP*AP*GP*TP*GP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*CP*AP*CP*TP*TP*CP*CP*GP*CP*TP*TP*AP*T)-3'), SODIUM ION, ... | | Authors: | Poon, G.M.K. | | Deposit date: | 2024-07-08 | | Release date: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Human PU.1 minimal ETS Domain (165-258) bound to d(AATAAGCGGAAGTGGG)
To Be Published
|
|
5A60
 
 | | Crystal structure of full-length E. coli ygiF in complex with tripolyphosphate and two magnesium ions | | Descriptor: | 1,2-ETHANEDIOL, INORGANIC TRIPHOSPHATASE, MAGNESIUM ION, ... | | Authors: | Martinez, J, Truffault, V, Hothorn, M. | | Deposit date: | 2015-06-23 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Determinants for Substrate Binding and Catalysis in Triphosphate Tunnel Metalloenzymes.
J.Biol.Chem., 290, 2015
|
|
