8XLV
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2.86 spike protein(6P), 1-RBD-up state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2023-12-26 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8XN5
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron EG.5.1 spike protein(6P) in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2023-12-29 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8XNF
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2.86 spike protein(6P) in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2023-12-29 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8Y18
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron JN.1 RBD in complex with human ACE2 (local refinement from the spike protein) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, L.J, Gu, Y.H, Qi, J.X, Gao, G.F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8Y5J
 
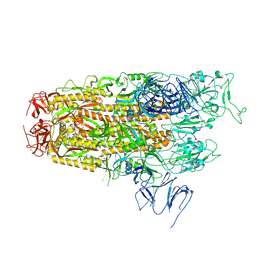 | | Cryo-EM structure of SARS-CoV-2 Omicron JN.1 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Li, L.J, Gu, Y.H, Qi, J.X, Gao, G.F. | | Deposit date: | 2024-01-31 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8Y16
 
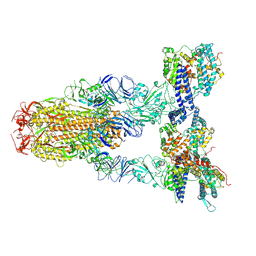 | | Cryo-EM structure of SARS-CoV-2 Omicron JN.1 spike protein in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, L.J, Gu, Y.H, Qi, J.X, Gao, G.F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8WZH
 
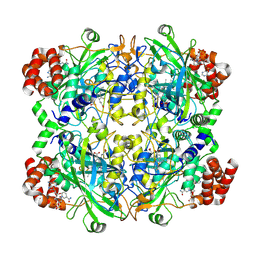 | |
8VI4
 
 | | TehA from Haemophilus influenzae purified in LMNG | | Descriptor: | Tellurite resistance protein TehA homolog | | Authors: | Catalano, C, Senko, S, Tran, N.L, Lucier, K.W, Farwell, A.C, Silva, M.S, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2024-01-03 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | High-Resolution Cryo-Electron Microscopy Structure Determination of Haemophilus influenzae Tellurite-Resistance Protein A via 200 kV Transmission Electron Microscopy.
Int J Mol Sci, 25, 2024
|
|
8UQM
 
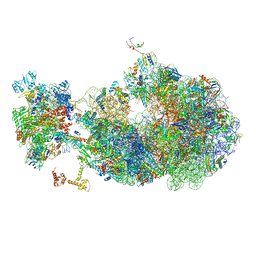 | | Escherichia coli transcription-translation coupled complex class B (TTC-B) containing RfaH in loaded state, NusA, mRNA with a 24 nt long spacer, and fMet-tRNAs in E-site and P-site of the ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Ebright, R.H. | | Deposit date: | 2023-10-24 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Structural basis of RfaH-mediated transcription-translation coupling.
Nat.Struct.Mol.Biol., 2024
|
|
8UPR
 
 | | Escherichia coli transcription-translation coupled complex class A (TTC-A) containing RfaH bound to ops signal, mRNA with a 21 nt long spacer, and fMet-tRNAs in E-site and P-site of the ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Ebright, R.H. | | Deposit date: | 2023-10-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Structural basis of RfaH-mediated transcription-translation coupling.
Nat.Struct.Mol.Biol., 2024
|
|
8UR0
 
 | | Escherichia coli transcription-translation coupled complex class B (TTC-B) containing RfaH bound to ops signal, NusA, mRNA with a 24 nt long spacer, and fMet-tRNAs in E-site and P-site of the ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Ebright, R.H. | | Deposit date: | 2023-10-25 | | Release date: | 2024-05-29 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of RfaH-mediated transcription-translation coupling.
Nat.Struct.Mol.Biol., 2024
|
|
8URX
 
 | | Escherichia coli transcription-translation coupled complex class B (TTC-B) containing RfaH bound to ops signal, mRNA with a 30 nt long spacer, and fMet-tRNAs in E-site and P-site of the ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Ebright, R.H. | | Deposit date: | 2023-10-27 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural basis of RfaH-mediated transcription-translation coupling.
Nat.Struct.Mol.Biol., 2024
|
|
8UPO
 
 | | Escherichia coli transcription-translation coupled complex class A (TTC-A) containing RfaH bound to ops signal, mRNA with a 21 nt long spacer, and fMet-tRNAs in E-site and P-site of the ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Ebright, R.H. | | Deposit date: | 2023-10-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structural basis of RfaH-mediated transcription-translation coupling.
Nat.Struct.Mol.Biol., 2024
|
|
7UPY
 
 | | An antibody from single human VH-rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Luo, S, Kreutzberger, A, Kirchhausen, T, Chen, B, Haynes, B, Alt, F. | | Deposit date: | 2022-04-18 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | An antibody from single human V H -rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion.
Sci Immunol, 7, 2022
|
|
8URI
 
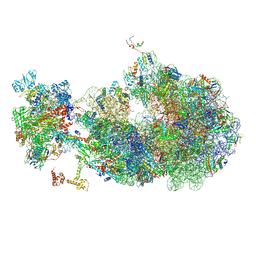 | | Escherichia coli transcription-translation coupled complex class B (TTC-B) containing RfaH bound to ops signal, NusA, mRNA with a 27 nt long spacer, and fMet-tRNAs in E-site and P-site of the ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Ebright, R.H. | | Deposit date: | 2023-10-26 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Structural basis of RfaH-mediated transcription-translation coupling.
Nat.Struct.Mol.Biol., 2024
|
|
8UDV
 
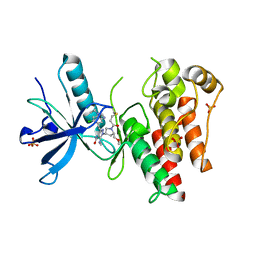 | | The X-RAY co-crystal structure of human FGFR3 V555M and Compound 17 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(6-chloro-1-cyclopropyl-1H-benzimidazol-5-yl)ethynyl]-1-[(3S,5S)-5-(methoxymethyl)-1-(prop-2-enoyl)pyrrolidin-3-yl]-5-(methylamino)-1H-pyrazole-4-carboxamide, Fibroblast growth factor receptor 3, ... | | Authors: | Tyhonas, J.S, Arnold, L.D, Cox, J, Franovic, A, Gardiner, E, Grandinetti, K, Kania, R, Kanouni, T, Lardy, M, Li, C, Martin, E.S, Miller, N, Mohan, A, Murphy, E.A, Perez, M, Soroceanu, L, Timple, N, Uryu, S, Womble, S, Kaldor, S.W. | | Deposit date: | 2023-09-29 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.348 Å) | | Cite: | Discovery of KIN-3248, An Irreversible, Next Generation FGFR Inhibitor for the Treatment of Advanced Tumors Harboring FGFR2 and/or FGFR3 Gene Alterations.
J.Med.Chem., 67, 2024
|
|
8VJJ
 
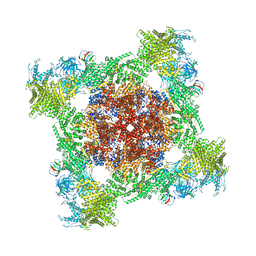 | | Structure of mouse RyR1 (EGTA-only dataset) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Peptidyl-prolyl cis-trans isomerase FKBP1A, Ryanodine receptor 1, ... | | Authors: | Weninger, G, Marks, A.R. | | Deposit date: | 2024-01-07 | | Release date: | 2024-01-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural insights into the regulation of RyR1 by S100A1.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7UPX
 
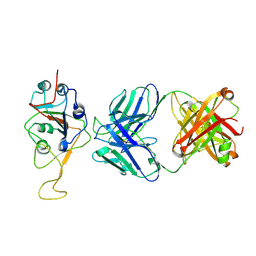 | | Three RBD-down state of SARS-CoV-2 D614G spike in complex with the SP1-77 neutralizing antibody Fab fragment (local refinement of the RBD and Fab variable domains) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, SP1-77 Fab heavy chain, SP1-77 Fab light chain, ... | | Authors: | Zhang, J, Luo, S, Kreutzberger, A, Kirchhausen, T, Chen, B, Haynes, B, Alt, F. | | Deposit date: | 2022-04-18 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | An antibody from single human V H -rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion.
Sci Immunol, 7, 2022
|
|
8VK4
 
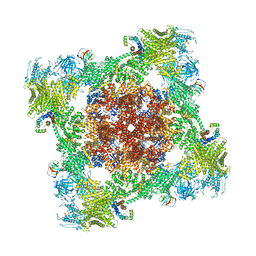 | |
8UZ1
 
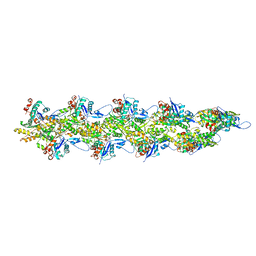 | | Straight actin filament from Arp2/3 branch junction sample (ADP-BeFx) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Chavali, S.S, Chou, S.Z, Sindelar, C.V. | | Deposit date: | 2023-11-14 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures reveal how phosphate release from Arp3 weakens actin filament branches formed by Arp2/3 complex.
Nat Commun, 15, 2024
|
|
7UPW
 
 | | Three RBD-down state of SARS-CoV-2 D614G spike in complex with the SP1-77 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Luo, S, Kreutzberger, A, Kirchhausen, T, Chen, B, Haynes, B, Alt, F. | | Deposit date: | 2022-04-18 | | Release date: | 2022-08-10 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | An antibody from single human V H -rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion.
Sci Immunol, 7, 2022
|
|
8UF7
 
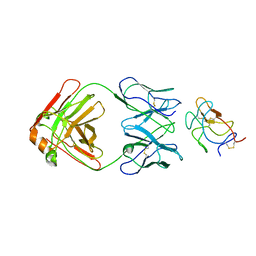 | | Cryo-EM structure of POmAb, a Type-I anti-prothrombin antiphospholipid antibody, bound to kringle-1 of human prothrombin | | Descriptor: | POmAb Heavy Chain, POmAb Light Chain, Prothrombin | | Authors: | Kumar, S, Summers, B, Basore, K, Pozzi, N. | | Deposit date: | 2023-10-03 | | Release date: | 2024-02-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure and functional basis of prothrombin recognition by a type I antiprothrombin antiphospholipid antibody.
Blood, 143, 2024
|
|
8VKK
 
 | | Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J, Poloni, C, Bezeruk, A, Tidey, K, Ahmed, S, Tuttle, K, Vahdatihassani, F, Cholak, S, Cook, L, Steiner, T.S, Subramaniam, S. | | Deposit date: | 2024-01-09 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | SARS-CoV-2 XBB.1.5 Spike Protein: Altered Receptor Binding, Antibody Evasion, and Retention of T Cell Recognition
To Be Published
|
|
8UXW
 
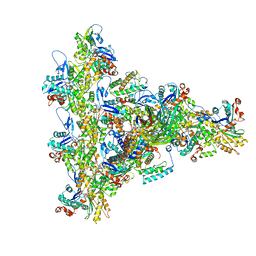 | | Arp2/3 branch junction complex, ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Chavali, S.S, Chou, S.Z, Sindelar, C.V. | | Deposit date: | 2023-11-11 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures reveal how phosphate release from Arp3 weakens actin filament branches formed by Arp2/3 complex.
Nat Commun, 15, 2024
|
|
8UDU
 
 | | The X-RAY co-crystal structure of human FGFR3 and Compound 17 | | Descriptor: | 3-[(6-chloro-1-cyclopropyl-1H-benzimidazol-5-yl)ethynyl]-1-[(3S,5S)-5-(methoxymethyl)-1-(prop-2-enoyl)pyrrolidin-3-yl]-5-(methylamino)-1H-pyrazole-4-carboxamide, CHLORIDE ION, Fibroblast growth factor receptor 3 | | Authors: | Tyhonas, J.S, Arnold, L.D, Cox, J, Franovic, A, Gardiner, E, Grandinetti, K, Kania, R, Kanouni, T, Lardy, M, Li, C, Martin, E.S, Miller, N, Mohan, A, Murphy, E.A, Perez, M, Soroceanu, L, Timple, N, Uryu, S, Womble, S, Kaldor, S.W. | | Deposit date: | 2023-09-29 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.737 Å) | | Cite: | Discovery of KIN-3248, An Irreversible, Next Generation FGFR Inhibitor for the Treatment of Advanced Tumors Harboring FGFR2 and/or FGFR3 Gene Alterations.
J.Med.Chem., 67, 2024
|
|
