3OGM
 
 | | Structure of COI1-ASK1 in complex with coronatine and the JAZ1 degron | | Descriptor: | (1S,2S)-2-ethyl-1-({[(3aS,4S,6R,7aS)-6-ethyl-1-oxooctahydro-1H-inden-4-yl]carbonyl}amino)cyclopropanecarboxylic acid, Coronatine-insensitive protein 1, JAZ1 degron peptide, ... | | Authors: | Sheard, L.B, Tan, X, Mao, H, Withers, J, Ben-Nissan, G, Hinds, T.R, Hsu, F, Sharon, M, Browse, J, He, S.Y, Rizo, J, Howe, G.A, Zheng, N. | | Deposit date: | 2010-08-17 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor.
Nature, 468, 2010
|
|
8EU3
 
 | | Cryo-EM structure of cGMP bound human CNGA3/CNGB3 channel in GDN, transition state 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYCLIC GUANOSINE MONOPHOSPHATE, Cyclic nucleotide-gated cation channel alpha-3, ... | | Authors: | Hu, Z, Zheng, X, Yang, J. | | Deposit date: | 2022-10-18 | | Release date: | 2023-08-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Conformational trajectory of allosteric gating of the human cone photoreceptor cyclic nucleotide-gated channel.
Nat Commun, 14, 2023
|
|
8ETP
 
 | | Cryo-EM structure of cGMP bound closed state of human CNGA3/CNGB3 channel in GDN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYCLIC GUANOSINE MONOPHOSPHATE, Cyclic nucleotide-gated cation channel alpha-3, ... | | Authors: | Hu, Z, Zheng, X, Yang, J. | | Deposit date: | 2022-10-17 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Conformational trajectory of allosteric gating of the human cone photoreceptor cyclic nucleotide-gated channel.
Nat Commun, 14, 2023
|
|
8EUC
 
 | | Cryo-EM structure of cGMP bound human CNGA3/CNGB3 channel in GDN, transition state 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYCLIC GUANOSINE MONOPHOSPHATE, Cyclic nucleotide-gated cation channel alpha-3, ... | | Authors: | Hu, Z, Zheng, X, Yang, J. | | Deposit date: | 2022-10-18 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Conformational trajectory of allosteric gating of the human cone photoreceptor cyclic nucleotide-gated channel.
Nat Commun, 14, 2023
|
|
3O6X
 
 | | Crystal Structure of the type III Glutamine Synthetase from Bacteroides fragilis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Glutamine synthetase, ... | | Authors: | van Rooyen, J.M, Belrhali, H, Abratt, V.R, Sewell, B.T. | | Deposit date: | 2010-07-29 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal Structure of Type III Glutamine Synthetase: Surprising Reversal of the Inter-Ring Interface.
Structure, 19, 2011
|
|
1LRQ
 
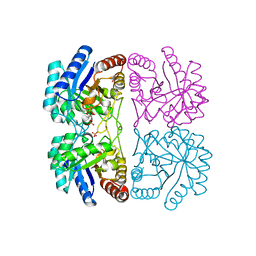 | | Aquifex aeolicus KDO8P synthase H185G mutant in complex with PEP, A5P and Cadmium | | Descriptor: | ARABINOSE-5-PHOSPHATE, CADMIUM ION, KDO-8-phosphate synthetase, ... | | Authors: | Wang, J, Duewel, H.S, Stuckey, J.A, Woodard, R.W, Gatti, D.L. | | Deposit date: | 2002-05-15 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Function of His185 in Aquifex aeolicus 3-Deoxy-D-manno-octulosonate 8-Phosphate Synthase
J.Mol.Biol., 324, 2002
|
|
3OGL
 
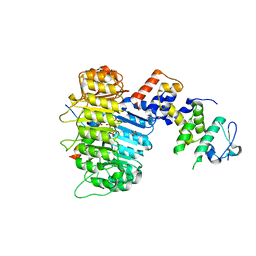 | | Structure of COI1-ASK1 in complex with JA-isoleucine and the JAZ1 degron | | Descriptor: | Coronatine-insensitive protein 1, JAZ1 incomplete degron peptide, N-({(1R,2S)-3-oxo-2-[(2Z)-pent-2-en-1-yl]cyclopentyl}acetyl)-L-isoleucine, ... | | Authors: | Sheard, L.B, Tan, X, Mao, H, Withers, J, Ben-Nissan, G, Hinds, T.R, Hsu, F, Sharon, M, Browse, J, He, S.Y, Rizo, J, Howe, G.A, Zheng, N. | | Deposit date: | 2010-08-17 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor.
Nature, 468, 2010
|
|
8JVE
 
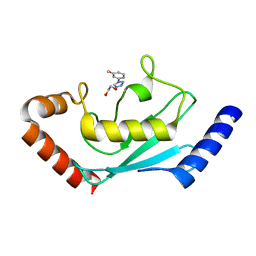 | | Identification and characterization of inhibitors covalently modifying catalytic cysteine of UBE2T and blocking ubiquitin transfer | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-methoxyphenyl)-1,2,3,4-tetrazole, Ubiquitin-conjugating enzyme E2 T | | Authors: | Anantharajan, J, Baburajendran, N. | | Deposit date: | 2023-06-28 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Identification and characterization of inhibitors covalently modifying catalytic cysteine of UBE2T and blocking ubiquitin transfer.
Biochem.Biophys.Res.Commun., 689, 2023
|
|
1AI3
 
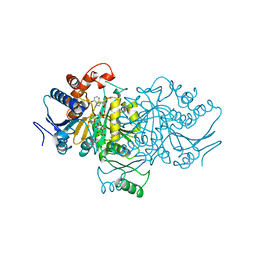 | | ORBITAL STEERING IN THE CATALYTIC POWER OF ENZYMES: SMALL STRUCTURAL CHANGES WITH LARGE CATALYTIC CONSEQUENCES | | Descriptor: | ISOCITRATE DEHYDROGENASE, ISOCITRIC ACID, MAGNESIUM ION, ... | | Authors: | Stoddard, B.L, Mesecar, A, Koshland Junior, D.E. | | Deposit date: | 1997-04-30 | | Release date: | 1997-11-12 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Orbital steering in the catalytic power of enzymes: small structural changes with large catalytic consequences.
Science, 277, 1997
|
|
8JVL
 
 | | Identification and characterization of inhibitors covalently modifying catalytic cysteine of UBE2T and blocking ubiquitin transfer | | Descriptor: | 1,2-ETHANEDIOL, 1-(4-methoxyphenyl)-1,2,3,4-tetrazole, Ubiquitin-conjugating enzyme E2 T | | Authors: | Anantharajan, J, Baburajendran, N. | | Deposit date: | 2023-06-28 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Identification and characterization of inhibitors covalently modifying catalytic cysteine of UBE2T and blocking ubiquitin transfer.
Biochem.Biophys.Res.Commun., 689, 2023
|
|
4YKI
 
 | |
1VZB
 
 | |
4YKJ
 
 | |
3KWG
 
 | |
4FYZ
 
 | | Crystal Structure of Nitrosyl Cytochrome P450cin | | Descriptor: | 1,3,3-TRIMETHYL-2-OXABICYCLO[2.2.2]OCTANE, DI(HYDROXYETHYL)ETHER, NITRIC OXIDE, ... | | Authors: | Madrona, Y, Tripathi, S.M, Li, H, Poulos, T.L. | | Deposit date: | 2012-07-05 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystal structures of substrate-free and nitrosyl cytochrome p450cin: implications for o(2) activation.
Biochemistry, 51, 2012
|
|
2W6Z
 
 | |
5FHT
 
 | | HtrA2 protease mutant V226K | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Golik, P, Dubin, G, Zurawa-Janicka, D, Lipinska, B, Jarzab, M, Wenta, T, Gieldon, A, Ciarkowski, A. | | Deposit date: | 2015-12-22 | | Release date: | 2016-09-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Distinct 3D Architecture and Dynamics of the Human HtrA2(Omi) Protease and Its Mutated Variants.
Plos One, 11, 2016
|
|
7PX5
 
 | | ATAD2 in complex with 1-Methyl-2-quinolone | | Descriptor: | 1,2-ETHANEDIOL, 1-METHYLQUINOLIN-2(1H)-ONE, ATPase family AAA domain-containing protein 2, ... | | Authors: | Martin, M.P, Noble, M.E.N. | | Deposit date: | 2021-10-07 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Exiting the tunnel of uncertainty: crystal soak to validated hit.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
8TG5
 
 | | tRNA 2'-phosphotransferase (Tpt1) from Pyrococcus horikoshii in complex with branched 2'-PO4 RNA | | Descriptor: | (2S,3R,4R,5S)-2-(6-amino-9H-purin-9-yl)-4-{[(S)-{[(2R,3S,4R,5S)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl]oxy}-5-({[(S)-{[(2R,3R,4S,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-2-(hydroxymethyl)oxolan-3-yl]oxy}(hydroxy)phosphoryl]oxy}methyl)oxolan-3-yl dihydrogen phosphate (non-preferred name), CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Jacewicz, A, Dantuluri, S, Shuman, S. | | Deposit date: | 2023-07-12 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for Tpt1-catalyzed 2'-PO 4 transfer from RNA and NADP(H) to NAD.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7MMZ
 
 | |
1DW1
 
 | | STRUCTURE OF THE CYANIDE COMPLEX OF SHP, AN OXYGEN BINDING CYTOCHROME C | | Descriptor: | CYANIDE ION, CYTOCHROME C, HEME C | | Authors: | Leys, D, Backers, K, Meyer, T.E, Hagen, W.R, Cusanovich, M.A, Van Beeumen, J.J. | | Deposit date: | 2000-01-24 | | Release date: | 2000-06-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of an oxygen-binding cytochrome c from Rhodobacter sphaeroides.
J.Biol.Chem., 275, 2000
|
|
1DW0
 
 | | STRUCTURE OF OXIDIZED SHP, AN OXYGEN BINDING CYTOCHROME C | | Descriptor: | CYTOCHROME C, HEME C, SULFATE ION | | Authors: | Leys, D, Backers, K, Meyer, T.E, Hagen, W.R, Cusanovich, M.A, Van Beeumen, J.J. | | Deposit date: | 2000-01-24 | | Release date: | 2000-06-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structures of an oxygen-binding cytochrome c from Rhodobacter sphaeroides.
J.Biol.Chem., 275, 2000
|
|
2W6O
 
 | | Crystal structure of Biotin carboxylase from E. coli in complex with 4-Amino-7,7-dimethyl-7,8-dihydro-quinazolinone fragment | | Descriptor: | 4-amino-7,7-dimethyl-7,8-dihydroquinazolin-5(6H)-one, BIOTIN CARBOXYLASE, CHLORIDE ION | | Authors: | Mochalkin, I, Miller, J.R. | | Deposit date: | 2008-12-18 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Antibacterial Biotin Carboxylase Inhibitors by Virtual Screening and Fragment-Based Approaches.
Acs Chem.Biol., 4, 2009
|
|
5EQ4
 
 | | Crystal structure of the SrpA adhesin R347E mutant from Streptococcus sanguinis | | Descriptor: | ACETATE ION, CALCIUM ION, Platelet-binding glycoprotein | | Authors: | Loukachevitch, L.V, McCulloch, K.M, Vann, K.R, Wawrzak, Z, Anderson, S, Iverson, T.M. | | Deposit date: | 2015-11-12 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Sialoglycan Binding by the Streptococcus sanguinis SrpA Adhesin.
J.Biol.Chem., 291, 2016
|
|
1DW3
 
 | | STRUCTURE OF A REDUCED OXYGEN BINDING CYTOCHROME C | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Leys, D, Backers, K, Meyer, T.E, Hagen, W.R, Cusanovich, M.A, Van Beeumen, J.J. | | Deposit date: | 2000-01-24 | | Release date: | 2000-06-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of an oxygen-binding cytochrome c from Rhodobacter sphaeroides.
J.Biol.Chem., 275, 2000
|
|
